Gene
KWMTBOMO06453 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011872
Annotation
PREDICTED:_calcium-transporting_ATPase_type_2C_member_1_isoform_X2_[Amyelois_transitella]
Full name
Calcium-transporting ATPase
Location in the cell
PlasmaMembrane Reliability : 3.907
Sequence
CDS
ATGTGGATCAGCGCAGTCGAGAGCTCTATACTGACATGGGAGGAGGTATCGGAGCGTTTGAGCGTCGACGTCCGCCGAGGCTTGTCATGGCGCGAAGCCAACGACCGCATGAACTTCGTTGGAGCCAACGAGTTTCAGGTGAAGGAACAAGATCCGCTCTGGAAGAAGTACATCGAGCAGTTCCAGAATCCGCTCATACTCCTGCTTCTCGGATCGGCGGTGGTGAGCGTTTGCATGCGCCAGTTCGACGACGCGATATCTATCACGGTCGCCATCATCATCGTGGTGACCGTCGCCTTCGTCCAGGAGTACCGCTCGGAGAAGTCGCTCGAGGAACTCAATAAACTTGTGCCTCCGTCATGCAATTGCCTTCGAGAGGGAAGTCTCGAGCACTTCCTCGCCCGCAACCTGGTCCCAGGCGACATCGTCCACCTGAACGTCGGGGACAGGGTGCCGGCCGACCTGCGGCTCTTCGACTCCACGGACCTGGCCATCGACGAGTCATCGTTCACCGGGGAAACAGAACCCGCCGTCAAGAACATCGTGCCAAACAAAGCGGCCGCCGGCAGGGTCAACAAGGATAACATAGCCTTCATGGGAACACTGGTTAGATGCGGAAATGCTAAGGGCATTGTGGTCAGCACGGGAGAACGTTCAGAGTTCGGTGACATTTTCCGCATGATGCAAGCCGAAGAAGCCCCGAAGACCCCGCTCCAGAAGTCCATGGATACCCTCGGGGGCCAGCTCTCGATGTACTCCTTCTGCATCATCGGCTTTATCATGATCACGGGGTGGCTCCAAGGAAAAGCCCTGCAGGAGATGTTCACTATCGGCGTCAGTCTCGCCGTTGCTGCCATACCCGAAGGTCTGCCAATAGTCGTGACTGTGACCCTGGCCCTCGGAGTCATGAGAATGGCGAAACGTAACGCCATAGTGAAGAAGCTGCCCACCGTTGAGACTTTGGGATGCGTTAATGTCATCTGCAGTGACAAGACGGGAACGTTGACGAAGAATGAAATGACAGTGACGACAATAGCGACATCCGGCGGCCACATCGCCGAAGTGACCGGAACGGGATACAACGGGCGCGGCGACATACAACTGCGAGCCTACAAGGGCAGAGATCTGGACGTGGCCAGGATGGGTGTCACCAGCATGTTGGAGGTGGCGGTACTGTGCAACAACGCGCACATCCGCGATGAGACGCTGTACGGGCAGCCCACGGAGGGCGCGCTCGTCGCCTGCGCCATGAAGCACGGCCTACGCGACCTCCGGGAACAGTACACCAGGCTGCACGAGATACCCTTCAGCTCAGAAACGAAGATGATGGTCGTGAAATGCGCCCCGAAGTTCTCCGACGGCGGCAAAGAGGAGTTGTTCGTGAAGGGGGCCGTCGAGAAGGTCCTGCCGCTGTGCACGCAGTACATCGACAGCGCCGGCAACTACGCGCCGCTCACGAACGAGAAGCAGAACGAAATATTAGCCGAGGTGTACAACATCGGACGAATGGGTTTAAGAACGATAGCTCTGTGCCGCGGCGCGAGCCTGGAGCGGCTGGTGTACGCGGGCGCTTGCGGCGTGTGCGACCCGCCGCGCCCCGGCGTGCGCGCCGCCGTGAGCACGCTGCGCCGCGCCGCCGTCGACGTCAAGATGGTGACCGGCGACGCGCGCCCCACGGCGCTCGCTGTCGCACAAATGGTCGGCTTGGACGTCCTGCACTCTCAATCTCTATCCGGCGAACAACTCGACTCGATGGCCGACGAAGAACTTGACCGCATTATCGATACGGTGAGCGTTTTCTACAGGGTGACGCCAAAGCACAAGCTTGCTATTGTGAAGTCTCTCCAGCGGCTCGGGAACATTGTCGGCATGACGGGTGACGGCGTCAACGACGGCGTGGCGCTGAAGCGGGCGGACATTGGCATTGCCATGGGCAAGAACGGCACCGACGTGTGCAAGGAGGCCGCCGACATGATCCTGGTCGATGATGACTTCGCCACCATCATAGCCGCAATCGAAGAAGGAAAATCGATATTCTACAACATTGGAAACTTCGTGAGGTTCCAACTGTCCACCTCGATAGCGGCGTTGTCGCTCATCGCTCTGGCAACACTGATGGGCGTTCCCAACCCGCTGAACGCTATGCAGATTCTCTGGATCAATATTATAATGGACGGTCCGCCGGCGCAGTCGCTGGGCGTGGAGCCCGTGGACCACGAGGTGGTGGGGCTGGGCCCGCGCGACACGCGGCGCCGCCTGCTGTCCCGCCCGCTGCTGCTCAACGTGCTGCTGTCCGCCGCCATCATCATCGCCGGCACGCTGTGGGTCTTCAATCGTGAGATGTCGGACAACACGATCACGCCCCGGGACACGACCATGACGTTCACGTGCTTCGTGCTGTTCGACATGTTCAACGCGCTGTCGTGCCGCTCGCAGACCAAGTCCGTGTTCGAGGTGGGGCTGCTCTCCAACCGCATGTTCCTGGCCGCCGTCACGCTGAGTCTGCTGGGCCAGGCCCTGGTTATTTACTTCCCGCCGTTGCAGAGCGTGTTCCAGACGGAGGCGCTCACAGCGCACGATATACTGTTCCTGGTCGGTCTTACGTCGAGCGTGTTTATAGTAAGCGAAATTAAAAAGCTTATCGAACGCACCCTGAAGAAGCGCGTCTACGGCAAGTACGCTGCTAAGGCCCACGACGCTATGGACTTTGTATGA
Protein
MWISAVESSILTWEEVSERLSVDVRRGLSWREANDRMNFVGANEFQVKEQDPLWKKYIEQFQNPLILLLLGSAVVSVCMRQFDDAISITVAIIIVVTVAFVQEYRSEKSLEELNKLVPPSCNCLREGSLEHFLARNLVPGDIVHLNVGDRVPADLRLFDSTDLAIDESSFTGETEPAVKNIVPNKAAAGRVNKDNIAFMGTLVRCGNAKGIVVSTGERSEFGDIFRMMQAEEAPKTPLQKSMDTLGGQLSMYSFCIIGFIMITGWLQGKALQEMFTIGVSLAVAAIPEGLPIVVTVTLALGVMRMAKRNAIVKKLPTVETLGCVNVICSDKTGTLTKNEMTVTTIATSGGHIAEVTGTGYNGRGDIQLRAYKGRDLDVARMGVTSMLEVAVLCNNAHIRDETLYGQPTEGALVACAMKHGLRDLREQYTRLHEIPFSSETKMMVVKCAPKFSDGGKEELFVKGAVEKVLPLCTQYIDSAGNYAPLTNEKQNEILAEVYNIGRMGLRTIALCRGASLERLVYAGACGVCDPPRPGVRAAVSTLRRAAVDVKMVTGDARPTALAVAQMVGLDVLHSQSLSGEQLDSMADEELDRIIDTVSVFYRVTPKHKLAIVKSLQRLGNIVGMTGDGVNDGVALKRADIGIAMGKNGTDVCKEAADMILVDDDFATIIAAIEEGKSIFYNIGNFVRFQLSTSIAALSLIALATLMGVPNPLNAMQILWINIIMDGPPAQSLGVEPVDHEVVGLGPRDTRRRLLSRPLLLNVLLSAAIIIAGTLWVFNREMSDNTITPRDTTMTFTCFVLFDMFNALSCRSQTKSVFEVGLLSNRMFLAAVTLSLLGQALVIYFPPLQSVFQTEALTAHDILFLVGLTSSVFIVSEIKKLIERTLKKRVYGKYAAKAHDAMDFV
Summary
Description
This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of calcium.
Catalytic Activity
ATP + Ca(2+)(in) + H2O = ADP + Ca(2+)(out) + H(+) + phosphate
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family.
Uniprot
A0A2A4JUZ5
A0A2A4JVP8
A0A3S2TKU5
A0A212ELX2
A0A2H1W7T5
A0A194PV50
+ More
A0A067RKU6 A0A2J7QLK3 A0A154PIL0 F4WDE3 A0A195BX72 A0A158NIS0 A0A151XDU7 A0A2A3E8J9 V9ICI9 A0A087ZVD3 A0A0J7N4A6 A0A195FTI2 A0A195CBY4 A0A0L7QSG7 E2BCM5 A0A0C9RCC7 E2AVH1 A0A151J7B8 A0A232F7A3 E0VUC0 A0A026W8F2 A0A0V0G3A0 A0A023F4S7 A0A224XJ47 A0A069DXQ9 A0A1B6ESV6 A0A1B6MH31 A0A1B6ML36 A0A1B6LFZ6 A0A1A9YUJ2 A0A0L0CEA4 A0A1I8MMU0 A0A2M4CUJ8 A0A1B6DL98 A0A2M4CT61 A0A1I8MMU8 A0A1B6EBU5 D6W6C6 A0A0M5JBF6 A0A1I8Q2K0 A0A1I8Q2J8 A0A1Q3F859 A0A1B0ESK7 A0A1Q3F7K6 A0A1Q3F825 A0A0K8TS12 A0A1S4F525 A0A0Q9XM47 A0A1Y1LA79 A0A1Y1L5I9 A0A0Q9XN45 A0A0Q9WWF4 A0A0Q9WSV2 A0A0Q9WKX5 A0A0Q5UK69 B4LDX9 A0A034VCB0 A0A0Q9WJU5 A0A0Q9XCR5 A0A0Q9XPK7 A0A0Q5UKR4 A0A0K8V5L8 B4KYJ6 A0A0Q9WJE3 A0A3B0JLL9 W8B6F5 A0A0R1E2R8 B3NEF6 W8BH99 A0A084WM47 A0A0Q5U613 A0A1W4UDS7 A0A1L8DF23 A0A1L8DEW2 A0A0R1E4G1 A0A182J722 A0A0R1E4B3 A0A1W4URA0 A0A0J9RZZ5 Q2M0Q6 C1C5A0 A0A1W4UR16 A0A0J9S025 A0A0J9S091 A0A0J9URQ8 Q9VNR2 B4PIQ1 A0A1B0DE25 B4IB41 Q8IPS6 Q7KTU2 B3MB86 A0A0P8YCD4 A0A1W4USS9
A0A067RKU6 A0A2J7QLK3 A0A154PIL0 F4WDE3 A0A195BX72 A0A158NIS0 A0A151XDU7 A0A2A3E8J9 V9ICI9 A0A087ZVD3 A0A0J7N4A6 A0A195FTI2 A0A195CBY4 A0A0L7QSG7 E2BCM5 A0A0C9RCC7 E2AVH1 A0A151J7B8 A0A232F7A3 E0VUC0 A0A026W8F2 A0A0V0G3A0 A0A023F4S7 A0A224XJ47 A0A069DXQ9 A0A1B6ESV6 A0A1B6MH31 A0A1B6ML36 A0A1B6LFZ6 A0A1A9YUJ2 A0A0L0CEA4 A0A1I8MMU0 A0A2M4CUJ8 A0A1B6DL98 A0A2M4CT61 A0A1I8MMU8 A0A1B6EBU5 D6W6C6 A0A0M5JBF6 A0A1I8Q2K0 A0A1I8Q2J8 A0A1Q3F859 A0A1B0ESK7 A0A1Q3F7K6 A0A1Q3F825 A0A0K8TS12 A0A1S4F525 A0A0Q9XM47 A0A1Y1LA79 A0A1Y1L5I9 A0A0Q9XN45 A0A0Q9WWF4 A0A0Q9WSV2 A0A0Q9WKX5 A0A0Q5UK69 B4LDX9 A0A034VCB0 A0A0Q9WJU5 A0A0Q9XCR5 A0A0Q9XPK7 A0A0Q5UKR4 A0A0K8V5L8 B4KYJ6 A0A0Q9WJE3 A0A3B0JLL9 W8B6F5 A0A0R1E2R8 B3NEF6 W8BH99 A0A084WM47 A0A0Q5U613 A0A1W4UDS7 A0A1L8DF23 A0A1L8DEW2 A0A0R1E4G1 A0A182J722 A0A0R1E4B3 A0A1W4URA0 A0A0J9RZZ5 Q2M0Q6 C1C5A0 A0A1W4UR16 A0A0J9S025 A0A0J9S091 A0A0J9URQ8 Q9VNR2 B4PIQ1 A0A1B0DE25 B4IB41 Q8IPS6 Q7KTU2 B3MB86 A0A0P8YCD4 A0A1W4USS9
EC Number
7.2.2.10
Pubmed
22118469
26354079
24845553
21719571
21347285
20798317
+ More
28648823 20566863 24508170 30249741 25474469 26334808 26108605 25315136 18362917 19820115 26369729 17994087 28004739 18057021 25348373 24495485 17550304 24438588 22936249 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
28648823 20566863 24508170 30249741 25474469 26334808 26108605 25315136 18362917 19820115 26369729 17994087 28004739 18057021 25348373 24495485 17550304 24438588 22936249 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
NWSH01000530
PCG75857.1
PCG75856.1
RSAL01000076
RVE48862.1
AGBW02013978
+ More
OWR42461.1 ODYU01006900 SOQ49165.1 KQ459590 KPI97197.1 KK852567 KDR21210.1 NEVH01013243 PNF29463.1 KQ434924 KZC11673.1 GL888087 EGI67723.1 KQ976394 KYM93239.1 ADTU01017107 ADTU01017108 KQ982289 KYQ58428.1 KZ288345 PBC27602.1 JR038699 AEY58372.1 LBMM01010365 KMQ87515.1 KQ981285 KYN43199.1 KQ978068 KYM97603.1 KQ414757 KOC61582.1 GL447336 EFN86535.1 GBYB01004853 GBYB01005960 JAG74620.1 JAG75727.1 GL443112 EFN62566.1 KQ979695 KYN19729.1 NNAY01000743 OXU26721.1 AAZO01005477 DS235785 EEB16976.1 KK107364 QOIP01000007 EZA51926.1 RLU20962.1 GECL01003530 JAP02594.1 GBBI01002290 JAC16422.1 GFTR01008263 JAW08163.1 GBGD01000397 JAC88492.1 GECZ01028684 GECZ01010224 JAS41085.1 JAS59545.1 GEBQ01004717 JAT35260.1 GEBQ01003353 JAT36624.1 GEBQ01017361 JAT22616.1 CCAG010019616 CCAG010019617 JRES01000502 KNC30576.1 GGFL01004340 MBW68518.1 GEDC01010834 JAS26464.1 GGFL01004339 MBW68517.1 GEDC01021944 GEDC01001897 JAS15354.1 JAS35401.1 KQ971319 EFA11368.2 CP012525 ALC44544.1 GFDL01011284 JAV23761.1 AJWK01004433 AJWK01004434 AJWK01004435 AJWK01004436 GFDL01011500 JAV23545.1 GFDL01011382 JAV23663.1 GDAI01000888 JAI16715.1 CH933809 KRG06372.1 GEZM01064051 JAV68845.1 GEZM01064052 JAV68844.1 KRG06366.1 CH940647 KRF84672.1 KRF84673.1 KRF84678.1 CH954178 KQS44438.1 EDW70022.1 KRF84677.1 GAKP01019235 GAKP01019234 JAC39717.1 KRF84674.1 KRG06374.1 KRG06375.1 KQS44439.1 GDHF01018143 GDHF01009762 JAI34171.1 JAI42552.1 EDW18807.1 KRF84675.1 OUUW01000002 SPP76290.1 GAMC01013942 GAMC01013941 JAB92613.1 CM000159 KRK02515.1 EDV52791.2 GAMC01013944 GAMC01013943 JAB92611.1 ATLV01024384 KE525351 KFB51291.1 KQS44437.1 GFDF01009099 JAV04985.1 GFDF01009098 JAV04986.1 KRK02516.1 KRK02514.1 CM002912 KMZ01227.1 CH379069 EAL30874.3 BT082029 AE014296 ACO72866.1 ACZ94767.1 KMZ01229.1 KMZ01228.1 KMZ01231.1 AAF51860.1 EDW95559.1 AJVK01014701 AJVK01014702 AJVK01014703 AJVK01014704 CH480826 EDW44504.1 BT133078 AAN12202.1 AEX93163.1 AAF51858.2 CH902618 EDV41387.2 KPU79086.1
OWR42461.1 ODYU01006900 SOQ49165.1 KQ459590 KPI97197.1 KK852567 KDR21210.1 NEVH01013243 PNF29463.1 KQ434924 KZC11673.1 GL888087 EGI67723.1 KQ976394 KYM93239.1 ADTU01017107 ADTU01017108 KQ982289 KYQ58428.1 KZ288345 PBC27602.1 JR038699 AEY58372.1 LBMM01010365 KMQ87515.1 KQ981285 KYN43199.1 KQ978068 KYM97603.1 KQ414757 KOC61582.1 GL447336 EFN86535.1 GBYB01004853 GBYB01005960 JAG74620.1 JAG75727.1 GL443112 EFN62566.1 KQ979695 KYN19729.1 NNAY01000743 OXU26721.1 AAZO01005477 DS235785 EEB16976.1 KK107364 QOIP01000007 EZA51926.1 RLU20962.1 GECL01003530 JAP02594.1 GBBI01002290 JAC16422.1 GFTR01008263 JAW08163.1 GBGD01000397 JAC88492.1 GECZ01028684 GECZ01010224 JAS41085.1 JAS59545.1 GEBQ01004717 JAT35260.1 GEBQ01003353 JAT36624.1 GEBQ01017361 JAT22616.1 CCAG010019616 CCAG010019617 JRES01000502 KNC30576.1 GGFL01004340 MBW68518.1 GEDC01010834 JAS26464.1 GGFL01004339 MBW68517.1 GEDC01021944 GEDC01001897 JAS15354.1 JAS35401.1 KQ971319 EFA11368.2 CP012525 ALC44544.1 GFDL01011284 JAV23761.1 AJWK01004433 AJWK01004434 AJWK01004435 AJWK01004436 GFDL01011500 JAV23545.1 GFDL01011382 JAV23663.1 GDAI01000888 JAI16715.1 CH933809 KRG06372.1 GEZM01064051 JAV68845.1 GEZM01064052 JAV68844.1 KRG06366.1 CH940647 KRF84672.1 KRF84673.1 KRF84678.1 CH954178 KQS44438.1 EDW70022.1 KRF84677.1 GAKP01019235 GAKP01019234 JAC39717.1 KRF84674.1 KRG06374.1 KRG06375.1 KQS44439.1 GDHF01018143 GDHF01009762 JAI34171.1 JAI42552.1 EDW18807.1 KRF84675.1 OUUW01000002 SPP76290.1 GAMC01013942 GAMC01013941 JAB92613.1 CM000159 KRK02515.1 EDV52791.2 GAMC01013944 GAMC01013943 JAB92611.1 ATLV01024384 KE525351 KFB51291.1 KQS44437.1 GFDF01009099 JAV04985.1 GFDF01009098 JAV04986.1 KRK02516.1 KRK02514.1 CM002912 KMZ01227.1 CH379069 EAL30874.3 BT082029 AE014296 ACO72866.1 ACZ94767.1 KMZ01229.1 KMZ01228.1 KMZ01231.1 AAF51860.1 EDW95559.1 AJVK01014701 AJVK01014702 AJVK01014703 AJVK01014704 CH480826 EDW44504.1 BT133078 AAN12202.1 AEX93163.1 AAF51858.2 CH902618 EDV41387.2 KPU79086.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000027135
UP000235965
+ More
UP000076502 UP000007755 UP000078540 UP000005205 UP000075809 UP000242457 UP000005203 UP000036403 UP000078541 UP000078542 UP000053825 UP000008237 UP000000311 UP000078492 UP000215335 UP000009046 UP000053097 UP000279307 UP000092444 UP000037069 UP000095301 UP000007266 UP000092553 UP000095300 UP000092461 UP000009192 UP000008792 UP000008711 UP000268350 UP000002282 UP000030765 UP000192221 UP000075880 UP000001819 UP000000803 UP000092462 UP000001292 UP000007801
UP000076502 UP000007755 UP000078540 UP000005205 UP000075809 UP000242457 UP000005203 UP000036403 UP000078541 UP000078542 UP000053825 UP000008237 UP000000311 UP000078492 UP000215335 UP000009046 UP000053097 UP000279307 UP000092444 UP000037069 UP000095301 UP000007266 UP000092553 UP000095300 UP000092461 UP000009192 UP000008792 UP000008711 UP000268350 UP000002282 UP000030765 UP000192221 UP000075880 UP000001819 UP000000803 UP000092462 UP000001292 UP000007801
Interpro
IPR023299
ATPase_P-typ_cyto_dom_N
+ More
IPR030336 ATP2C1
IPR006413 P-type_ATPase_IIA_PMR1
IPR023298 ATPase_P-typ_TM_dom_sf
IPR006068 ATPase_P-typ_cation-transptr_C
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR001757 P_typ_ATPase
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR018303 ATPase_P-typ_P_site
IPR004014 ATPase_P-typ_cation-transptr_N
IPR030334 ATP2C2
IPR030336 ATP2C1
IPR006413 P-type_ATPase_IIA_PMR1
IPR023298 ATPase_P-typ_TM_dom_sf
IPR006068 ATPase_P-typ_cation-transptr_C
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR001757 P_typ_ATPase
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR018303 ATPase_P-typ_P_site
IPR004014 ATPase_P-typ_cation-transptr_N
IPR030334 ATP2C2
Gene 3D
ProteinModelPortal
A0A2A4JUZ5
A0A2A4JVP8
A0A3S2TKU5
A0A212ELX2
A0A2H1W7T5
A0A194PV50
+ More
A0A067RKU6 A0A2J7QLK3 A0A154PIL0 F4WDE3 A0A195BX72 A0A158NIS0 A0A151XDU7 A0A2A3E8J9 V9ICI9 A0A087ZVD3 A0A0J7N4A6 A0A195FTI2 A0A195CBY4 A0A0L7QSG7 E2BCM5 A0A0C9RCC7 E2AVH1 A0A151J7B8 A0A232F7A3 E0VUC0 A0A026W8F2 A0A0V0G3A0 A0A023F4S7 A0A224XJ47 A0A069DXQ9 A0A1B6ESV6 A0A1B6MH31 A0A1B6ML36 A0A1B6LFZ6 A0A1A9YUJ2 A0A0L0CEA4 A0A1I8MMU0 A0A2M4CUJ8 A0A1B6DL98 A0A2M4CT61 A0A1I8MMU8 A0A1B6EBU5 D6W6C6 A0A0M5JBF6 A0A1I8Q2K0 A0A1I8Q2J8 A0A1Q3F859 A0A1B0ESK7 A0A1Q3F7K6 A0A1Q3F825 A0A0K8TS12 A0A1S4F525 A0A0Q9XM47 A0A1Y1LA79 A0A1Y1L5I9 A0A0Q9XN45 A0A0Q9WWF4 A0A0Q9WSV2 A0A0Q9WKX5 A0A0Q5UK69 B4LDX9 A0A034VCB0 A0A0Q9WJU5 A0A0Q9XCR5 A0A0Q9XPK7 A0A0Q5UKR4 A0A0K8V5L8 B4KYJ6 A0A0Q9WJE3 A0A3B0JLL9 W8B6F5 A0A0R1E2R8 B3NEF6 W8BH99 A0A084WM47 A0A0Q5U613 A0A1W4UDS7 A0A1L8DF23 A0A1L8DEW2 A0A0R1E4G1 A0A182J722 A0A0R1E4B3 A0A1W4URA0 A0A0J9RZZ5 Q2M0Q6 C1C5A0 A0A1W4UR16 A0A0J9S025 A0A0J9S091 A0A0J9URQ8 Q9VNR2 B4PIQ1 A0A1B0DE25 B4IB41 Q8IPS6 Q7KTU2 B3MB86 A0A0P8YCD4 A0A1W4USS9
A0A067RKU6 A0A2J7QLK3 A0A154PIL0 F4WDE3 A0A195BX72 A0A158NIS0 A0A151XDU7 A0A2A3E8J9 V9ICI9 A0A087ZVD3 A0A0J7N4A6 A0A195FTI2 A0A195CBY4 A0A0L7QSG7 E2BCM5 A0A0C9RCC7 E2AVH1 A0A151J7B8 A0A232F7A3 E0VUC0 A0A026W8F2 A0A0V0G3A0 A0A023F4S7 A0A224XJ47 A0A069DXQ9 A0A1B6ESV6 A0A1B6MH31 A0A1B6ML36 A0A1B6LFZ6 A0A1A9YUJ2 A0A0L0CEA4 A0A1I8MMU0 A0A2M4CUJ8 A0A1B6DL98 A0A2M4CT61 A0A1I8MMU8 A0A1B6EBU5 D6W6C6 A0A0M5JBF6 A0A1I8Q2K0 A0A1I8Q2J8 A0A1Q3F859 A0A1B0ESK7 A0A1Q3F7K6 A0A1Q3F825 A0A0K8TS12 A0A1S4F525 A0A0Q9XM47 A0A1Y1LA79 A0A1Y1L5I9 A0A0Q9XN45 A0A0Q9WWF4 A0A0Q9WSV2 A0A0Q9WKX5 A0A0Q5UK69 B4LDX9 A0A034VCB0 A0A0Q9WJU5 A0A0Q9XCR5 A0A0Q9XPK7 A0A0Q5UKR4 A0A0K8V5L8 B4KYJ6 A0A0Q9WJE3 A0A3B0JLL9 W8B6F5 A0A0R1E2R8 B3NEF6 W8BH99 A0A084WM47 A0A0Q5U613 A0A1W4UDS7 A0A1L8DF23 A0A1L8DEW2 A0A0R1E4G1 A0A182J722 A0A0R1E4B3 A0A1W4URA0 A0A0J9RZZ5 Q2M0Q6 C1C5A0 A0A1W4UR16 A0A0J9S025 A0A0J9S091 A0A0J9URQ8 Q9VNR2 B4PIQ1 A0A1B0DE25 B4IB41 Q8IPS6 Q7KTU2 B3MB86 A0A0P8YCD4 A0A1W4USS9
PDB
6HEF
E-value=8.62043e-106,
Score=984
Ontologies
GO
PANTHER
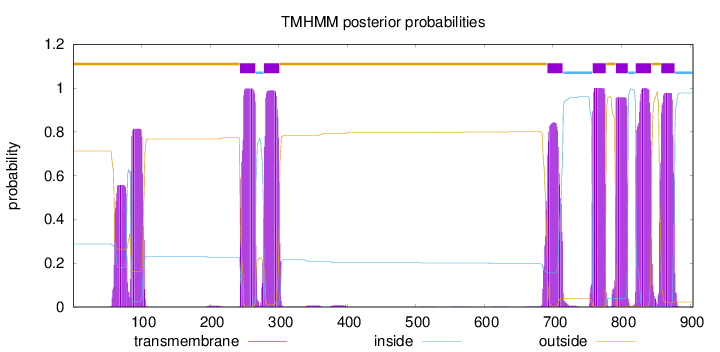
Topology
Subcellular location
Membrane
Length:
904
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
170.05112
Exp number, first 60 AAs:
0.40755
Total prob of N-in:
0.28775
outside
1 - 243
TMhelix
244 - 266
inside
267 - 278
TMhelix
279 - 301
outside
302 - 691
TMhelix
692 - 714
inside
715 - 757
TMhelix
758 - 777
outside
778 - 791
TMhelix
792 - 809
inside
810 - 820
TMhelix
821 - 843
outside
844 - 857
TMhelix
858 - 877
inside
878 - 904
Population Genetic Test Statistics
Pi
249.279445
Theta
168.927373
Tajima's D
0.647142
CLR
1.312067
CSRT
0.559622018899055
Interpretation
Uncertain
