Gene
KWMTBOMO06451
Pre Gene Modal
BGIBMGA011609
Annotation
beta-1?3-glucan_recognition_protein_2_precursor_[Bombyx_mori]
Full name
Beta-1,3-glucan-binding protein
+ More
Beta-1,3-glucan-binding protein 1
Beta-1,3-glucan-binding protein 1
Alternative Name
Beta-1,3-glucan recognition protein
Gram negative bacteria-binding protein
Beta-1,3-glucan recognition protein 1
Gram negative bacteria-binding protein
Beta-1,3-glucan recognition protein 1
Location in the cell
Cytoplasmic Reliability : 1.04 Extracellular Reliability : 1.035 Nuclear Reliability : 1.025
Sequence
CDS
ATGACACTGTTCGCGTTCCAAGGCAACTTGAACCATAAACTGGATAGCACCAGCGTCGGGACACTGAGTGCAGAGGTACTGGATCCAGTCAACGGCCGATGGGTGTACGAAGAGCCCGACCTTAAACTAAAAGTCAAGGACGTCGTCTATTATAACGCGGTCTTCTCAATCAACAAGAAATTATACGAGAAAACAAACCAACAGTTCACCATAACAGAGCTAGAAGATCCTAATGCAAGCACAGATTCTCAGAAACCAGAATACAATATTCGAGGAGCAATTTGA
Protein
MTLFAFQGNLNHKLDSTSVGTLSAEVLDPVNGRWVYEEPDLKLKVKDVVYYNAVFSINKKLYEKTNQQFTITELEDPNASTDSQKPEYNIRGAI
Summary
Description
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and activates the phenoloxidase cascade (By similarity).
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and activates the phenoloxidase cascade.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan, causing aggregation of invading micro-organisms and activation of the phenoloxidase cascade.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and activates the phenoloxidase cascade. Causes aggregation of invading micro-organisms.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and activates the phenoloxidase cascade.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan, causing aggregation of invading micro-organisms and activation of the phenoloxidase cascade.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and activates the phenoloxidase cascade. Causes aggregation of invading micro-organisms.
Subunit
Monomer.
Similarity
Belongs to the insect beta-1,3-glucan binding protein family.
Keywords
Glycoprotein
Immunity
Innate immunity
Secreted
Signal
Direct protein sequencing
3D-structure
Complete proteome
Reference proteome
Feature
chain Beta-1,3-glucan-binding protein
Uniprot
H9JQ04
Q17233
A0A3G1VCG4
E7D1Z3
V9XSA5
T1VXX4
+ More
A0A2A4JVZ1 A0A2H1WU49 B7SVL5 A0A2W1BUK8 A0A212ELV5 I4DP06 O96363 A0A194PUV7 A0A0N1IPG2 I4DN17 A0A2J7QLN9 A0A2J7QLS7 A0A067RND0 A0A1W4XSN8 A0A1Y1LR95 A0A0K8UT21 W8BJK5 A0A1W4VG67 A0A034VYC5 A0A346RAD2 H8YU83 A0A0K8W7M8 B4MKF8 V5GXH2 A0A0A1XKC9 B4PAK5 A1ZBU5 G2J5W3 B3NJK3 Q28Z72 B4J5K6 A0ZX45 A0A0L0C2S5 A0ZX43 B3MFX0 A0A1L8E2T0 Q76DI2 K7JGU7 Q8MU95 A0A0M4EW55 B4QF07 A0A1Y1MGE0 A0A3S2NJ43 B4J3L1 A0A0P8XM40 A0A0D3L473 B4GI71 A0A3B0J1U3 A0A1B6EH17 B1B5K2 A0A232ES71 A0A1A9V4J2 A0A194PW19 A0A2A4JH28 H8YU81 A0A212ESM0 C7AHP9 S4NX93 B4KPI9 A0A1B6EG16 A0A0A1WR53 B4KPJ0 B4P2U9 D6WIF9 A0A336MP99 A0A1W4XH40 C7AHP8 A0A0N0PD03 A0A2H1WUA9 B4HQ78 A0A2W1BWP9 Q9NJ98 M9ZRM8 H9JQ03 B3N8N2 A0A0Q9WZJ0 A0A1Y1LC59 A0A194Q1C0 Q9NL89 B3MHJ9 B3NBL7 A0A1B6E6G9 B4MMC4 A0A087ZVU9 A0A0N1PIG9
A0A2A4JVZ1 A0A2H1WU49 B7SVL5 A0A2W1BUK8 A0A212ELV5 I4DP06 O96363 A0A194PUV7 A0A0N1IPG2 I4DN17 A0A2J7QLN9 A0A2J7QLS7 A0A067RND0 A0A1W4XSN8 A0A1Y1LR95 A0A0K8UT21 W8BJK5 A0A1W4VG67 A0A034VYC5 A0A346RAD2 H8YU83 A0A0K8W7M8 B4MKF8 V5GXH2 A0A0A1XKC9 B4PAK5 A1ZBU5 G2J5W3 B3NJK3 Q28Z72 B4J5K6 A0ZX45 A0A0L0C2S5 A0ZX43 B3MFX0 A0A1L8E2T0 Q76DI2 K7JGU7 Q8MU95 A0A0M4EW55 B4QF07 A0A1Y1MGE0 A0A3S2NJ43 B4J3L1 A0A0P8XM40 A0A0D3L473 B4GI71 A0A3B0J1U3 A0A1B6EH17 B1B5K2 A0A232ES71 A0A1A9V4J2 A0A194PW19 A0A2A4JH28 H8YU81 A0A212ESM0 C7AHP9 S4NX93 B4KPI9 A0A1B6EG16 A0A0A1WR53 B4KPJ0 B4P2U9 D6WIF9 A0A336MP99 A0A1W4XH40 C7AHP8 A0A0N0PD03 A0A2H1WUA9 B4HQ78 A0A2W1BWP9 Q9NJ98 M9ZRM8 H9JQ03 B3N8N2 A0A0Q9WZJ0 A0A1Y1LC59 A0A194Q1C0 Q9NL89 B3MHJ9 B3NBL7 A0A1B6E6G9 B4MMC4 A0A087ZVU9 A0A0N1PIG9
Pubmed
19121390
8755572
27989837
21296155
19033442
28756777
+ More
22118469 22651552 9818384 26354079 24845553 28004739 24495485 25348373 29910977 17994087 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17103056 26108605 12923175 20075255 12770576 22936249 18195005 28648823 19387012 23622113 18362917 19820115 10713054 14976985 23497397 10671539 3136171
22118469 22651552 9818384 26354079 24845553 28004739 24495485 25348373 29910977 17994087 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17103056 26108605 12923175 20075255 12770576 22936249 18195005 28648823 19387012 23622113 18362917 19820115 10713054 14976985 23497397 10671539 3136171
EMBL
BABH01012373
L38591
AAB40946.1
MG732914
AYK02796.1
HQ007028
+ More
ADT82662.1 KF725771 AHD25001.1 KF425324 AGT95925.1 NWSH01000530 PCG75858.1 ODYU01011077 SOQ56588.1 EU770384 ACI32828.1 KZ149896 PZC78762.1 AGBW02013978 OWR42458.1 AK403230 BAM19646.1 AF023916 AAD09290.1 KQ459590 KPI97191.1 KQ460391 KPJ15396.1 AK402685 BAM19307.1 NEVH01013243 PNF29497.1 PNF29498.1 KK852567 KDR21214.1 GEZM01054584 JAV73547.1 GDHF01022809 JAI29505.1 GAMC01016766 JAB89789.1 GAKP01011860 JAC47092.1 MH200922 AXS59132.1 JF915525 AFD54027.1 GDHF01005232 JAI47082.1 CH963846 EDW72597.1 GALX01002004 JAB66462.1 GBXI01002972 JAD11320.1 CM000158 EDW91396.1 AE013599 AAF57465.1 BT128820 AEM72516.1 CH954179 EDV55310.1 CM000071 EAL25742.2 CH916367 EDW00769.1 AM050221 AM050225 AM050226 AM050227 AM050229 CAJ18893.1 CAJ18897.1 CAJ18898.1 CAJ18899.1 CAJ18901.1 JRES01001077 KNC25719.1 AM050219 AM050220 AM050222 AM050223 AM050224 AM050228 AM050230 CAJ18891.1 CAJ18892.1 CAJ18894.1 CAJ18895.1 CAJ18896.1 CAJ18900.1 CAJ18902.1 CH902619 EDV35652.2 GFDF01001056 JAV13028.1 AB108841 BAC99308.1 AF532603 AAM95970.1 CP012524 ALC42250.1 CM000362 CM002911 EDX07918.1 KMY95269.1 GEZM01031711 JAV84859.1 RSAL01000076 RVE48860.1 CH916366 EDV96213.1 KPU75777.1 JQ218446 AFU52688.1 CH479183 EDW36191.1 OUUW01000001 SPP72902.1 GEDC01000083 JAS37215.1 AB363981 BAG14263.1 NNAY01002525 OXU21136.1 KPI97193.1 NWSH01001422 PCG71281.1 JF915523 AFD54025.1 AGBW02012766 OWR44496.1 FJ546104 ACT66881.1 GAIX01012252 JAA80308.1 CH933808 EDW10185.1 GEDC01000412 JAS36886.1 GBXI01013287 JAD01005.1 EDW10186.2 CM000157 EDW89360.1 KQ971334 EFA01356.1 UFQS01001336 UFQT01001336 SSX10401.1 SSX30087.1 FJ546103 ACT66880.1 KPJ15397.1 SOQ56587.1 CH480816 EDW48703.1 PZC78761.1 AF177982 AAF44011.1 KC355197 AGK40897.1 CH954177 EDV59509.1 KRF97601.1 GEZM01062254 JAV69920.1 KPI97195.1 AB026441 BAA92243.1 EDV35835.1 CH954178 EDV50613.1 GEDC01003809 JAS33489.1 CH963847 EDW73269.1 KPJ15398.1
ADT82662.1 KF725771 AHD25001.1 KF425324 AGT95925.1 NWSH01000530 PCG75858.1 ODYU01011077 SOQ56588.1 EU770384 ACI32828.1 KZ149896 PZC78762.1 AGBW02013978 OWR42458.1 AK403230 BAM19646.1 AF023916 AAD09290.1 KQ459590 KPI97191.1 KQ460391 KPJ15396.1 AK402685 BAM19307.1 NEVH01013243 PNF29497.1 PNF29498.1 KK852567 KDR21214.1 GEZM01054584 JAV73547.1 GDHF01022809 JAI29505.1 GAMC01016766 JAB89789.1 GAKP01011860 JAC47092.1 MH200922 AXS59132.1 JF915525 AFD54027.1 GDHF01005232 JAI47082.1 CH963846 EDW72597.1 GALX01002004 JAB66462.1 GBXI01002972 JAD11320.1 CM000158 EDW91396.1 AE013599 AAF57465.1 BT128820 AEM72516.1 CH954179 EDV55310.1 CM000071 EAL25742.2 CH916367 EDW00769.1 AM050221 AM050225 AM050226 AM050227 AM050229 CAJ18893.1 CAJ18897.1 CAJ18898.1 CAJ18899.1 CAJ18901.1 JRES01001077 KNC25719.1 AM050219 AM050220 AM050222 AM050223 AM050224 AM050228 AM050230 CAJ18891.1 CAJ18892.1 CAJ18894.1 CAJ18895.1 CAJ18896.1 CAJ18900.1 CAJ18902.1 CH902619 EDV35652.2 GFDF01001056 JAV13028.1 AB108841 BAC99308.1 AF532603 AAM95970.1 CP012524 ALC42250.1 CM000362 CM002911 EDX07918.1 KMY95269.1 GEZM01031711 JAV84859.1 RSAL01000076 RVE48860.1 CH916366 EDV96213.1 KPU75777.1 JQ218446 AFU52688.1 CH479183 EDW36191.1 OUUW01000001 SPP72902.1 GEDC01000083 JAS37215.1 AB363981 BAG14263.1 NNAY01002525 OXU21136.1 KPI97193.1 NWSH01001422 PCG71281.1 JF915523 AFD54025.1 AGBW02012766 OWR44496.1 FJ546104 ACT66881.1 GAIX01012252 JAA80308.1 CH933808 EDW10185.1 GEDC01000412 JAS36886.1 GBXI01013287 JAD01005.1 EDW10186.2 CM000157 EDW89360.1 KQ971334 EFA01356.1 UFQS01001336 UFQT01001336 SSX10401.1 SSX30087.1 FJ546103 ACT66880.1 KPJ15397.1 SOQ56587.1 CH480816 EDW48703.1 PZC78761.1 AF177982 AAF44011.1 KC355197 AGK40897.1 CH954177 EDV59509.1 KRF97601.1 GEZM01062254 JAV69920.1 KPI97195.1 AB026441 BAA92243.1 EDV35835.1 CH954178 EDV50613.1 GEDC01003809 JAS33489.1 CH963847 EDW73269.1 KPJ15398.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000235965
+ More
UP000027135 UP000192223 UP000192221 UP000007798 UP000002282 UP000000803 UP000008711 UP000001819 UP000001070 UP000037069 UP000007801 UP000002358 UP000092553 UP000000304 UP000283053 UP000008744 UP000268350 UP000215335 UP000078200 UP000009192 UP000007266 UP000001292 UP000005203
UP000027135 UP000192223 UP000192221 UP000007798 UP000002282 UP000000803 UP000008711 UP000001819 UP000001070 UP000037069 UP000007801 UP000002358 UP000092553 UP000000304 UP000283053 UP000008744 UP000268350 UP000215335 UP000078200 UP000009192 UP000007266 UP000001292 UP000005203
Interpro
SUPFAM
SSF49899
SSF49899
Gene 3D
ProteinModelPortal
H9JQ04
Q17233
A0A3G1VCG4
E7D1Z3
V9XSA5
T1VXX4
+ More
A0A2A4JVZ1 A0A2H1WU49 B7SVL5 A0A2W1BUK8 A0A212ELV5 I4DP06 O96363 A0A194PUV7 A0A0N1IPG2 I4DN17 A0A2J7QLN9 A0A2J7QLS7 A0A067RND0 A0A1W4XSN8 A0A1Y1LR95 A0A0K8UT21 W8BJK5 A0A1W4VG67 A0A034VYC5 A0A346RAD2 H8YU83 A0A0K8W7M8 B4MKF8 V5GXH2 A0A0A1XKC9 B4PAK5 A1ZBU5 G2J5W3 B3NJK3 Q28Z72 B4J5K6 A0ZX45 A0A0L0C2S5 A0ZX43 B3MFX0 A0A1L8E2T0 Q76DI2 K7JGU7 Q8MU95 A0A0M4EW55 B4QF07 A0A1Y1MGE0 A0A3S2NJ43 B4J3L1 A0A0P8XM40 A0A0D3L473 B4GI71 A0A3B0J1U3 A0A1B6EH17 B1B5K2 A0A232ES71 A0A1A9V4J2 A0A194PW19 A0A2A4JH28 H8YU81 A0A212ESM0 C7AHP9 S4NX93 B4KPI9 A0A1B6EG16 A0A0A1WR53 B4KPJ0 B4P2U9 D6WIF9 A0A336MP99 A0A1W4XH40 C7AHP8 A0A0N0PD03 A0A2H1WUA9 B4HQ78 A0A2W1BWP9 Q9NJ98 M9ZRM8 H9JQ03 B3N8N2 A0A0Q9WZJ0 A0A1Y1LC59 A0A194Q1C0 Q9NL89 B3MHJ9 B3NBL7 A0A1B6E6G9 B4MMC4 A0A087ZVU9 A0A0N1PIG9
A0A2A4JVZ1 A0A2H1WU49 B7SVL5 A0A2W1BUK8 A0A212ELV5 I4DP06 O96363 A0A194PUV7 A0A0N1IPG2 I4DN17 A0A2J7QLN9 A0A2J7QLS7 A0A067RND0 A0A1W4XSN8 A0A1Y1LR95 A0A0K8UT21 W8BJK5 A0A1W4VG67 A0A034VYC5 A0A346RAD2 H8YU83 A0A0K8W7M8 B4MKF8 V5GXH2 A0A0A1XKC9 B4PAK5 A1ZBU5 G2J5W3 B3NJK3 Q28Z72 B4J5K6 A0ZX45 A0A0L0C2S5 A0ZX43 B3MFX0 A0A1L8E2T0 Q76DI2 K7JGU7 Q8MU95 A0A0M4EW55 B4QF07 A0A1Y1MGE0 A0A3S2NJ43 B4J3L1 A0A0P8XM40 A0A0D3L473 B4GI71 A0A3B0J1U3 A0A1B6EH17 B1B5K2 A0A232ES71 A0A1A9V4J2 A0A194PW19 A0A2A4JH28 H8YU81 A0A212ESM0 C7AHP9 S4NX93 B4KPI9 A0A1B6EG16 A0A0A1WR53 B4KPJ0 B4P2U9 D6WIF9 A0A336MP99 A0A1W4XH40 C7AHP8 A0A0N0PD03 A0A2H1WUA9 B4HQ78 A0A2W1BWP9 Q9NJ98 M9ZRM8 H9JQ03 B3N8N2 A0A0Q9WZJ0 A0A1Y1LC59 A0A194Q1C0 Q9NL89 B3MHJ9 B3NBL7 A0A1B6E6G9 B4MMC4 A0A087ZVU9 A0A0N1PIG9
PDB
2KHA
E-value=4.07054e-08,
Score=131
Ontologies
GO
PANTHER
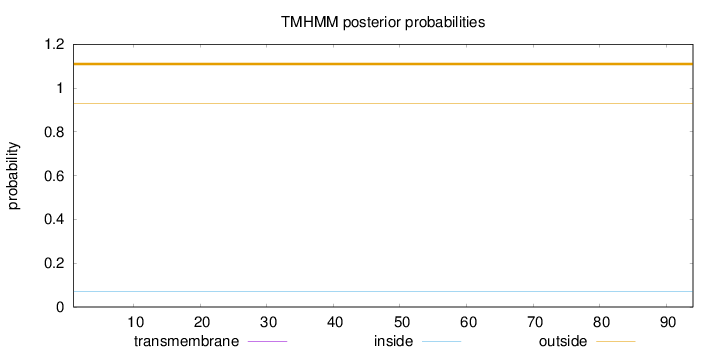
Topology
Subcellular location
Secreted
Length:
94
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00015
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.06938
outside
1 - 94
Population Genetic Test Statistics
Pi
153.055256
Theta
134.100512
Tajima's D
-1.302328
CLR
535.921895
CSRT
0.0894955252237388
Interpretation
Uncertain
