Gene
KWMTBOMO06450 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011608
Annotation
beta-1?3-glucan_recognition_protein_1_[Helicoverpa_armigera]
Full name
Beta-1,3-glucan-binding protein
+ More
Beta-1,3-glucan-binding protein 1
Beta-1,3-glucan-binding protein 2
Gram-negative bacteria-binding protein 3
Beta-1,3-glucan-binding protein 1
Beta-1,3-glucan-binding protein 2
Gram-negative bacteria-binding protein 3
Alternative Name
Beta-1,3-glucan recognition protein
Beta-1,3-glucan recognition protein 1
Beta-1,3-glucan recognition protein 2
Beta-1,3-glucan recognition protein 1
Beta-1,3-glucan recognition protein 2
Location in the cell
Cytoplasmic Reliability : 2.385
Sequence
CDS
ATGTACAAAACATGTGTGTGGGTCTTGTTATTTAAAATCGTGCTGTGCTACGAGGCACCACCGGCCACGCTCGAAGCAATACACCCTAAAGGACTTAGGGTTTCCGTTCCAGATGAGGGCTTCTCGTTATTCGCGTTTCACGGTAAACTCAACGAGGAAATGGAAGGCTTAGAGGCCGGCCATTGGTCCAGGGACATCACGAAGCCAAAAAACGGAAGATGGATATTCAGAGATCGAAATGCTGCGCTGAAAGTCGGAGATAAGATTTACTTTTGGACTTTTGTCATAAAGGACGGCTTAGGATACAGACAGGATAACGGGGAGTGGACAGTTGAAGGTTTCGTAGATGAAGCCGGTAATCCAGTAAACACAGAGGGCTCTGAAATAACACCAGGAGTAGAATTCACCAGCACCAGTCTGAACCCAGAGTCCCCTCAGTCTATACCCAATCAGCCTCCAGACAGCCTGCCCGCTAAACCACCATCTGAAGGCTATCCCTGTGAGCTATCGCTATCCACAGTCTCAGTACCTGGTTTTGTATGCAAAGGGCAGCTGCTTTTCGAAGATCAGTTCAATATACCAATACACAAAGGGAAAATTTGGGTACCAGAAGTAAAGTTCCCTGGTGAACCGGATTTTCCCTTCAACGTATACCTAACCGATAATGCAGAAGTCAACGACGGAAGGCTGATTATAAAACCAACTACCTTGGAGTCAAAGTATGGCGAGGATTTTGTTCGACAATCTTTGGATCTGTCAGAGAGATGCACTGGAACCGTAGGAACAGCTCAATGCTTGCGAGAAGCTTCTGGTCCTCTTATTTTGCCACCCATAATAACTGCGAAAATCAGCACTCGGCATCAATTTGCCTTCAAATATGGACGTGTCGAGATCAGAGCTAAAATGCCAAAAGGAGATTGGCTTTATCCTGAAATATTGCTCGAACCTCGTGATAATATATATGGTGTCCGGAACTACGCTTCGGGGATATTAAAAATCGCTAGCGTCAAAGGAAACGCTGAATTTTCAAAGAAACTCTACGCAGGTCCTATAATGACTGGATCTGATCCTTATAGATCTTTTTATCTTAAAGAAAACATTGGGTACGAATCATGGAATAACGATTTCCACAACTACACCCTAGAGTGGAGACCAGATGGCATTACGTTGTTAGTTGATGGAGAAAGTTATGGCGAGATAAAACCAGGAGAAGGTTTTTACAACGTGGCCAACTCATATAAAGTTGAAGCAGCGCCGCAGTGGCTTAAAGGCACAATCATGGCGCCTTTTGATGAATTGTTCTATGTGTCGATCGGTCTTAATGTGGCTGGAATCCGAGAGTTCTCCGAAGACATCTCCAATAAGCCTTGGAAAAACTCTGCCACTAAAGCAATGCTCAAATTCTGGGACGCCCGTAGTCAGTGGTTCCCTACGTGGGATGAAGATAGCGCTCTTCAGGTGGACTATGTTAAAGTTTTTGCGATATGA
Protein
MYKTCVWVLLFKIVLCYEAPPATLEAIHPKGLRVSVPDEGFSLFAFHGKLNEEMEGLEAGHWSRDITKPKNGRWIFRDRNAALKVGDKIYFWTFVIKDGLGYRQDNGEWTVEGFVDEAGNPVNTEGSEITPGVEFTSTSLNPESPQSIPNQPPDSLPAKPPSEGYPCELSLSTVSVPGFVCKGQLLFEDQFNIPIHKGKIWVPEVKFPGEPDFPFNVYLTDNAEVNDGRLIIKPTTLESKYGEDFVRQSLDLSERCTGTVGTAQCLREASGPLILPPIITAKISTRHQFAFKYGRVEIRAKMPKGDWLYPEILLEPRDNIYGVRNYASGILKIASVKGNAEFSKKLYAGPIMTGSDPYRSFYLKENIGYESWNNDFHNYTLEWRPDGITLLVDGESYGEIKPGEGFYNVANSYKVEAAPQWLKGTIMAPFDELFYVSIGLNVAGIREFSEDISNKPWKNSATKAMLKFWDARSQWFPTWDEDSALQVDYVKVFAI
Summary
Description
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and activates the phenoloxidase cascade.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and activates the phenoloxidase cascade. Causes aggregation of invading micro-organisms.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan, causing aggregation of invading micro-organisms and activation of the phenoloxidase cascade.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and lipoteichoic acid and causes aggregation of invading micro-organisms. Binding to beta-1,3-glucan activates the phenoloxidase cascade.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and activates the phenoloxidase cascade (By similarity).
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and activates the phenoloxidase cascade. Causes aggregation of invading micro-organisms.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan, causing aggregation of invading micro-organisms and activation of the phenoloxidase cascade.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and lipoteichoic acid and causes aggregation of invading micro-organisms. Binding to beta-1,3-glucan activates the phenoloxidase cascade.
Involved in the recognition of invading micro-organisms. Binds specifically to beta-1,3-glucan and activates the phenoloxidase cascade (By similarity).
Subunit
Monomer.
Similarity
Belongs to the insect beta-1,3-glucan binding protein family.
Keywords
3D-structure
Complete proteome
Direct protein sequencing
Glycoprotein
Immunity
Innate immunity
Reference proteome
Secreted
Signal
Feature
chain Beta-1,3-glucan-binding protein
Uniprot
H9JQ03
Q9NL89
A0A2H1WUA9
A0A2W1BWP9
B7SVL2
A0A2A4JH28
+ More
Q0E666 A0A194PUV7 A0A212ELU9 I3RJG9 Q9NJ98 B7SVK8 A0A0N0PD03 G3MVG5 A0A0D3L473 A0A2A4JGS8 A0A2W1BZR7 B5BSX8 A0A194Q1C0 A0A0N1PIG9 Q8MU95 Q8ISB6 H9AA72 H9JQ02 A0A346RAD2 A0A194PW19 A0A212ESM0 A0A3S2L9V1 S4NX93 B7SVK9 F2YLD9 B7SVL3 A0A0N1IPP2 A0A194PUW2 A0A2H1WSV4 A0A194PWQ7 I4DP05 I4DNK4 A0A0L7LKY4 B7SVK4 A0A2A4JIQ0 B7SVK6 G5DE93 B7SVL4 A0A2W1C0X2 G3MVG4 A0A2H1WUD7 D6WIF9 A0A2J7QLS7 B4KZH1 B4QMM7 B3M5X3 B3NBL7 A0A3B0JVG9 Q76DI2 B4MMC4 A8E788 Q9NHA8 B5X538 B4HK50 B4PFH1 A0A1Y1JZ37 B4LEW9 A0A1W4XH40 J3JUM4 Q29DZ2 A0A1Y1JZ48 A0A067RND0 A0A1W4XSN8 B4J3L1 A0A1W4VCK9 N6TB38 U4UDM5 Q17NM6 A0A1Y1K3B6 A0A0L0C2S5 A0A1I8NZT4 H8YU83 A0A1Y9IW14 A0A182GFY0 A0A1S4H971 A0A3F2Z1Y8 A0A1C7CZK4 A0A182HQD5 A0A3F2YXP9 A0A182MYU8 W5JFT8 A0A182P5V2 H8YU81 A0A3F2YU17 A0A182MML6 A0A1I8M2R2 A0A0K8UT21 A0A3F2YX59 Q7Q0E5 A0A0C9PSC7
Q0E666 A0A194PUV7 A0A212ELU9 I3RJG9 Q9NJ98 B7SVK8 A0A0N0PD03 G3MVG5 A0A0D3L473 A0A2A4JGS8 A0A2W1BZR7 B5BSX8 A0A194Q1C0 A0A0N1PIG9 Q8MU95 Q8ISB6 H9AA72 H9JQ02 A0A346RAD2 A0A194PW19 A0A212ESM0 A0A3S2L9V1 S4NX93 B7SVK9 F2YLD9 B7SVL3 A0A0N1IPP2 A0A194PUW2 A0A2H1WSV4 A0A194PWQ7 I4DP05 I4DNK4 A0A0L7LKY4 B7SVK4 A0A2A4JIQ0 B7SVK6 G5DE93 B7SVL4 A0A2W1C0X2 G3MVG4 A0A2H1WUD7 D6WIF9 A0A2J7QLS7 B4KZH1 B4QMM7 B3M5X3 B3NBL7 A0A3B0JVG9 Q76DI2 B4MMC4 A8E788 Q9NHA8 B5X538 B4HK50 B4PFH1 A0A1Y1JZ37 B4LEW9 A0A1W4XH40 J3JUM4 Q29DZ2 A0A1Y1JZ48 A0A067RND0 A0A1W4XSN8 B4J3L1 A0A1W4VCK9 N6TB38 U4UDM5 Q17NM6 A0A1Y1K3B6 A0A0L0C2S5 A0A1I8NZT4 H8YU83 A0A1Y9IW14 A0A182GFY0 A0A1S4H971 A0A3F2Z1Y8 A0A1C7CZK4 A0A182HQD5 A0A3F2YXP9 A0A182MYU8 W5JFT8 A0A182P5V2 H8YU81 A0A3F2YU17 A0A182MML6 A0A1I8M2R2 A0A0K8UT21 A0A3F2YX59 Q7Q0E5 A0A0C9PSC7
Pubmed
19121390
10671539
3136171
28756777
19033442
26354079
+ More
22118469 10713054 14976985 21910994 18840462 12770576 29910977 23622113 22651552 26227816 18362917 19820115 17994087 22936249 12923175 10827089 17103056 10731132 12537572 17550304 28004739 22516182 15632085 24845553 23537049 17510324 26108605 26483478 12364791 20966253 20920257 23761445 25315136
22118469 10713054 14976985 21910994 18840462 12770576 29910977 23622113 22651552 26227816 18362917 19820115 17994087 22936249 12923175 10827089 17103056 10731132 12537572 17550304 28004739 22516182 15632085 24845553 23537049 17510324 26108605 26483478 12364791 20966253 20920257 23761445 25315136
EMBL
BABH01012373
AB026441
BAA92243.1
ODYU01011077
SOQ56587.1
KZ149896
+ More
PZC78761.1 EU770381 ACI32825.1 NWSH01001422 PCG71281.1 AM265582 CAK22401.1 KQ459590 KPI97191.1 AGBW02013978 OWR42456.1 JQ692072 AFK24449.1 AF177982 AAF44011.1 EU770377 ACI32821.1 KQ460391 KPJ15397.1 HM459596 ADZ45541.1 JQ218446 AFU52688.1 PCG71285.1 PZC78757.1 AB436166 BAG70413.1 KPI97195.1 KPJ15398.1 AF532603 AAM95970.1 AY135522 AAN10151.1 JN880424 AFC35297.1 MH200922 AXS59132.1 KPI97193.1 AGBW02012766 OWR44496.1 RSAL01000076 RVE48856.1 GAIX01012252 JAA80308.1 EU770378 ACI32822.1 JF313110 ADZ93428.1 EU770382 ACI32826.1 KPJ15399.1 KPI97196.1 ODYU01010749 SOQ56056.1 KPI97194.1 AK403229 BAM19645.1 AK402982 BAM19494.1 JTDY01000692 KOB76198.1 EU770373 ACI32817.1 PCG71282.1 EU770375 ACI32819.1 JN641797 AEQ33590.1 EU770383 ACI32827.1 PZC78760.1 HM459595 ADZ45540.2 SOQ56586.1 KQ971334 EFA01356.1 NEVH01013243 PNF29498.1 CH933809 EDW17898.1 CM000363 CM002912 EDX09781.1 KMY98515.1 CH902618 EDV39663.1 CH954178 EDV50613.1 OUUW01000002 SPP76731.1 AB108841 BAC99308.1 CH963847 EDW73269.1 BT031030 ABV82412.1 AF228474 AM050231 AM050232 AM050233 AM050234 AM050235 AM050236 AM050237 AM050238 AM050239 AM050240 AM050241 AM050242 AE014296 AAF33851.1 AAF50349.1 BT046157 ACI46545.1 CH480815 EDW40786.1 CM000159 EDW93097.1 GEZM01096755 JAV54582.1 CH940647 EDW69138.1 BT126939 AEE61901.1 CH379070 EAL30271.1 GEZM01096759 JAV54579.1 KK852567 KDR21214.1 CH916366 EDV96213.1 APGK01044526 APGK01044527 KB741026 ENN74953.1 KB632309 ERL92074.1 CH477198 EAT48288.2 GEZM01096757 JAV54580.1 JRES01001077 KNC25719.1 JF915525 AFD54027.1 JXUM01060643 KQ562111 KXJ76671.1 AAAB01008986 APCN01003309 ADMH02001279 ETN63242.1 JF915523 AFD54025.1 AXCM01006744 GDHF01022809 JAI29505.1 AXCN02001026 EAA00167.3 GBYB01004253 JAG74020.1
PZC78761.1 EU770381 ACI32825.1 NWSH01001422 PCG71281.1 AM265582 CAK22401.1 KQ459590 KPI97191.1 AGBW02013978 OWR42456.1 JQ692072 AFK24449.1 AF177982 AAF44011.1 EU770377 ACI32821.1 KQ460391 KPJ15397.1 HM459596 ADZ45541.1 JQ218446 AFU52688.1 PCG71285.1 PZC78757.1 AB436166 BAG70413.1 KPI97195.1 KPJ15398.1 AF532603 AAM95970.1 AY135522 AAN10151.1 JN880424 AFC35297.1 MH200922 AXS59132.1 KPI97193.1 AGBW02012766 OWR44496.1 RSAL01000076 RVE48856.1 GAIX01012252 JAA80308.1 EU770378 ACI32822.1 JF313110 ADZ93428.1 EU770382 ACI32826.1 KPJ15399.1 KPI97196.1 ODYU01010749 SOQ56056.1 KPI97194.1 AK403229 BAM19645.1 AK402982 BAM19494.1 JTDY01000692 KOB76198.1 EU770373 ACI32817.1 PCG71282.1 EU770375 ACI32819.1 JN641797 AEQ33590.1 EU770383 ACI32827.1 PZC78760.1 HM459595 ADZ45540.2 SOQ56586.1 KQ971334 EFA01356.1 NEVH01013243 PNF29498.1 CH933809 EDW17898.1 CM000363 CM002912 EDX09781.1 KMY98515.1 CH902618 EDV39663.1 CH954178 EDV50613.1 OUUW01000002 SPP76731.1 AB108841 BAC99308.1 CH963847 EDW73269.1 BT031030 ABV82412.1 AF228474 AM050231 AM050232 AM050233 AM050234 AM050235 AM050236 AM050237 AM050238 AM050239 AM050240 AM050241 AM050242 AE014296 AAF33851.1 AAF50349.1 BT046157 ACI46545.1 CH480815 EDW40786.1 CM000159 EDW93097.1 GEZM01096755 JAV54582.1 CH940647 EDW69138.1 BT126939 AEE61901.1 CH379070 EAL30271.1 GEZM01096759 JAV54579.1 KK852567 KDR21214.1 CH916366 EDV96213.1 APGK01044526 APGK01044527 KB741026 ENN74953.1 KB632309 ERL92074.1 CH477198 EAT48288.2 GEZM01096757 JAV54580.1 JRES01001077 KNC25719.1 JF915525 AFD54027.1 JXUM01060643 KQ562111 KXJ76671.1 AAAB01008986 APCN01003309 ADMH02001279 ETN63242.1 JF915523 AFD54025.1 AXCM01006744 GDHF01022809 JAI29505.1 AXCN02001026 EAA00167.3 GBYB01004253 JAG74020.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000283053
+ More
UP000037510 UP000007266 UP000235965 UP000009192 UP000000304 UP000007801 UP000008711 UP000268350 UP000007798 UP000000803 UP000001292 UP000002282 UP000008792 UP000192223 UP000001819 UP000027135 UP000001070 UP000192221 UP000019118 UP000030742 UP000008820 UP000037069 UP000095300 UP000075920 UP000069940 UP000249989 UP000076407 UP000075882 UP000075840 UP000075900 UP000075884 UP000000673 UP000075885 UP000075881 UP000075883 UP000095301 UP000075886 UP000007062
UP000037510 UP000007266 UP000235965 UP000009192 UP000000304 UP000007801 UP000008711 UP000268350 UP000007798 UP000000803 UP000001292 UP000002282 UP000008792 UP000192223 UP000001819 UP000027135 UP000001070 UP000192221 UP000019118 UP000030742 UP000008820 UP000037069 UP000095300 UP000075920 UP000069940 UP000249989 UP000076407 UP000075882 UP000075840 UP000075900 UP000075884 UP000000673 UP000075885 UP000075881 UP000075883 UP000095301 UP000075886 UP000007062
Interpro
SUPFAM
SSF49899
SSF49899
Gene 3D
ProteinModelPortal
H9JQ03
Q9NL89
A0A2H1WUA9
A0A2W1BWP9
B7SVL2
A0A2A4JH28
+ More
Q0E666 A0A194PUV7 A0A212ELU9 I3RJG9 Q9NJ98 B7SVK8 A0A0N0PD03 G3MVG5 A0A0D3L473 A0A2A4JGS8 A0A2W1BZR7 B5BSX8 A0A194Q1C0 A0A0N1PIG9 Q8MU95 Q8ISB6 H9AA72 H9JQ02 A0A346RAD2 A0A194PW19 A0A212ESM0 A0A3S2L9V1 S4NX93 B7SVK9 F2YLD9 B7SVL3 A0A0N1IPP2 A0A194PUW2 A0A2H1WSV4 A0A194PWQ7 I4DP05 I4DNK4 A0A0L7LKY4 B7SVK4 A0A2A4JIQ0 B7SVK6 G5DE93 B7SVL4 A0A2W1C0X2 G3MVG4 A0A2H1WUD7 D6WIF9 A0A2J7QLS7 B4KZH1 B4QMM7 B3M5X3 B3NBL7 A0A3B0JVG9 Q76DI2 B4MMC4 A8E788 Q9NHA8 B5X538 B4HK50 B4PFH1 A0A1Y1JZ37 B4LEW9 A0A1W4XH40 J3JUM4 Q29DZ2 A0A1Y1JZ48 A0A067RND0 A0A1W4XSN8 B4J3L1 A0A1W4VCK9 N6TB38 U4UDM5 Q17NM6 A0A1Y1K3B6 A0A0L0C2S5 A0A1I8NZT4 H8YU83 A0A1Y9IW14 A0A182GFY0 A0A1S4H971 A0A3F2Z1Y8 A0A1C7CZK4 A0A182HQD5 A0A3F2YXP9 A0A182MYU8 W5JFT8 A0A182P5V2 H8YU81 A0A3F2YU17 A0A182MML6 A0A1I8M2R2 A0A0K8UT21 A0A3F2YX59 Q7Q0E5 A0A0C9PSC7
Q0E666 A0A194PUV7 A0A212ELU9 I3RJG9 Q9NJ98 B7SVK8 A0A0N0PD03 G3MVG5 A0A0D3L473 A0A2A4JGS8 A0A2W1BZR7 B5BSX8 A0A194Q1C0 A0A0N1PIG9 Q8MU95 Q8ISB6 H9AA72 H9JQ02 A0A346RAD2 A0A194PW19 A0A212ESM0 A0A3S2L9V1 S4NX93 B7SVK9 F2YLD9 B7SVL3 A0A0N1IPP2 A0A194PUW2 A0A2H1WSV4 A0A194PWQ7 I4DP05 I4DNK4 A0A0L7LKY4 B7SVK4 A0A2A4JIQ0 B7SVK6 G5DE93 B7SVL4 A0A2W1C0X2 G3MVG4 A0A2H1WUD7 D6WIF9 A0A2J7QLS7 B4KZH1 B4QMM7 B3M5X3 B3NBL7 A0A3B0JVG9 Q76DI2 B4MMC4 A8E788 Q9NHA8 B5X538 B4HK50 B4PFH1 A0A1Y1JZ37 B4LEW9 A0A1W4XH40 J3JUM4 Q29DZ2 A0A1Y1JZ48 A0A067RND0 A0A1W4XSN8 B4J3L1 A0A1W4VCK9 N6TB38 U4UDM5 Q17NM6 A0A1Y1K3B6 A0A0L0C2S5 A0A1I8NZT4 H8YU83 A0A1Y9IW14 A0A182GFY0 A0A1S4H971 A0A3F2Z1Y8 A0A1C7CZK4 A0A182HQD5 A0A3F2YXP9 A0A182MYU8 W5JFT8 A0A182P5V2 H8YU81 A0A3F2YU17 A0A182MML6 A0A1I8M2R2 A0A0K8UT21 A0A3F2YX59 Q7Q0E5 A0A0C9PSC7
PDB
3AQX
E-value=6.9861e-56,
Score=551
Ontologies
GO
PANTHER
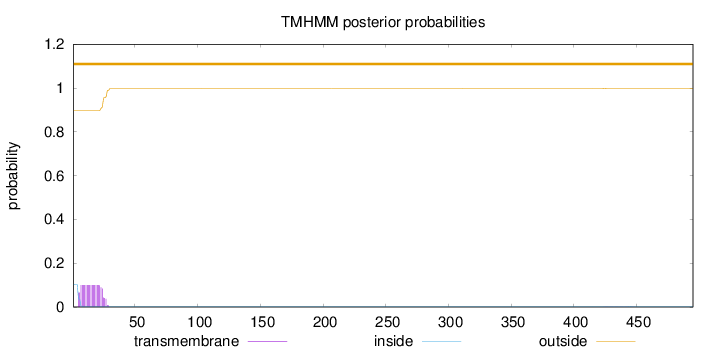
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 16,
Likelihood: 0.982657

Length:
495
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.06287
Exp number, first 60 AAs:
2.04647
Total prob of N-in:
0.10275
outside
1 - 495
Population Genetic Test Statistics
Pi
239.896865
Theta
166.067419
Tajima's D
-1.358286
CLR
1122.481897
CSRT
0.079646017699115
Interpretation
Uncertain
