Gene
KWMTBOMO06443
Pre Gene Modal
BGIBMGA011876
Annotation
PREDICTED:_elongation_of_very_long_chain_fatty_acids_protein_7-like_isoform_X2_[Bombyx_mori]
Full name
Elongation of very long chain fatty acids protein
+ More
Elongation of very long chain fatty acids protein 7
Elongation of very long chain fatty acids protein 7
Alternative Name
Very-long-chain 3-oxoacyl-CoA synthase
3-keto acyl-CoA synthase ELOVL7
ELOVL fatty acid elongase 7
Very long chain 3-ketoacyl-CoA synthase 7
Very long chain 3-oxoacyl-CoA synthase 7
3-keto acyl-CoA synthase ELOVL7
ELOVL fatty acid elongase 7
Very long chain 3-ketoacyl-CoA synthase 7
Very long chain 3-oxoacyl-CoA synthase 7
Location in the cell
PlasmaMembrane Reliability : 2.838
Sequence
CDS
ATGGAAAGAATAGATGTGCCATCGTTCTGGGAGTTCAAAGGAGCCGGTGATTATACGGATCAAAAACTATTGATGTCATCCCCATTTCCTGTTACCACGATTATAATCCTATACTTGTGGTTCGTTTTGAAATTCGGTCCGGAATTCATGAGGCAACGAAAACCCTACAATGTTGAAAAGCTTCTTGTTGTATACAACAATTTACAAGTTGCCGTCTCGTTTTACATCTTTTATTTGGAAAAATCGTAA
Protein
MERIDVPSFWEFKGAGDYTDQKLLMSSPFPVTTIIILYLWFVLKFGPEFMRQRKPYNVEKLLVVYNNLQVAVSFYIFYLEKS
Summary
Description
Catalyzes the first and rate-limiting reaction of the four reactions that constitute the long-chain fatty acids elongation cycle. This endoplasmic reticulum-bound enzymatic process allows the addition of 2 carbons to the chain of long- and very long-chain fatty acids (VLCFAs) per cycle. Condensing enzyme with higher activity toward C18 acyl-CoAs, especially C18:3(n-3) acyl-CoAs and C18:3(n-6)-CoAs. Also active toward C20:4-, C18:0-, C18:1-, C18:2- and C16:0-CoAs, and weakly toward C20:0-CoA. Little or no activity toward C22:0-, C24:0-, or C26:0-CoAs. May participate to the production of saturated and polyunsaturated VLCFAs of different chain lengths that are involved in multiple biological processes as precursors of membrane lipids and lipid mediators.
Catalytic Activity
a very-long-chain acyl-CoA + H(+) + malonyl-CoA = a very-long-chain 3-oxoacyl-CoA + CO2 + CoA
Similarity
Belongs to the ELO family.
Belongs to the ELO family. ELOVL7 subfamily.
Belongs to the ELO family. ELOVL7 subfamily.
Uniprot
H9JQS0
A0A0L7LJB9
A0A2H1VQJ9
A0A2H1WH44
A0A1E1WBM7
A0A0K8VDZ8
+ More
A0A0K8VS01 A0A0A1XAS6 A0A1A9XJ22 A0A1B0BXZ2 A0A034W2T7 W8AWR6 A0A1B0BXZ1 A0A1A9XJ24 A0A034WG82 A0A0K8VXE8 A0A1A9V757 A0A1A9Z493 A0A1A9V750 A0A1A9Z486 A0A182VTN0 A0A1B0FQK1 A0A1B0FQL7 A0A194PYL9 A0A0M4F374 A0A2R7WMF5 A0A1A9W5Z6 A0A182RA82 A0A182M983 A0A1B6CDH2 T1PEI5 Q17HH4 A0A182T880 A0A182X629 A0A182VEV3 A0A182TLQ9 A0A182LJX9 Q7PPN4 A0A182PW57 A0A3B0JEF4 A0A182F0V7 A0A182IB52 A0A182K6I1 A0A182Y8I0 A0A084VMS9 A0A182QAU9 A0A212EWH4 A0A023EP01 Q29BK4 W5J4Z1 A0A182ISK0 B4GP37 B4NJS5 A0A182NB63 A0A1Q3G529 A0A0A1XAY8 A0A1B0D2D9 A0A0L0C049 W8BNS1 A0A3P8ZCE6 A0A0L7RD74 A0A1I8NVN9 A0A1W4WV09 A0A1A9W5Z8 A0A1W4WV14 A0A2A3EJY7 E9ICJ1 A0A3B0JJ32 A0A0L0CGV6 A0A2D0R3G6 B4M165 E3TCK8 J9JX82 A0A1I8M3I9 A0A146M9A3 B4K958 B4GP38 Q29BK5 A0A067R7E0 A0A3P8XC63 A0A1A8H4U3 A0A087ZPK7 B3M000 A0A2J7QVP9 A0A1E1W7T1
A0A0K8VS01 A0A0A1XAS6 A0A1A9XJ22 A0A1B0BXZ2 A0A034W2T7 W8AWR6 A0A1B0BXZ1 A0A1A9XJ24 A0A034WG82 A0A0K8VXE8 A0A1A9V757 A0A1A9Z493 A0A1A9V750 A0A1A9Z486 A0A182VTN0 A0A1B0FQK1 A0A1B0FQL7 A0A194PYL9 A0A0M4F374 A0A2R7WMF5 A0A1A9W5Z6 A0A182RA82 A0A182M983 A0A1B6CDH2 T1PEI5 Q17HH4 A0A182T880 A0A182X629 A0A182VEV3 A0A182TLQ9 A0A182LJX9 Q7PPN4 A0A182PW57 A0A3B0JEF4 A0A182F0V7 A0A182IB52 A0A182K6I1 A0A182Y8I0 A0A084VMS9 A0A182QAU9 A0A212EWH4 A0A023EP01 Q29BK4 W5J4Z1 A0A182ISK0 B4GP37 B4NJS5 A0A182NB63 A0A1Q3G529 A0A0A1XAY8 A0A1B0D2D9 A0A0L0C049 W8BNS1 A0A3P8ZCE6 A0A0L7RD74 A0A1I8NVN9 A0A1W4WV09 A0A1A9W5Z8 A0A1W4WV14 A0A2A3EJY7 E9ICJ1 A0A3B0JJ32 A0A0L0CGV6 A0A2D0R3G6 B4M165 E3TCK8 J9JX82 A0A1I8M3I9 A0A146M9A3 B4K958 B4GP38 Q29BK5 A0A067R7E0 A0A3P8XC63 A0A1A8H4U3 A0A087ZPK7 B3M000 A0A2J7QVP9 A0A1E1W7T1
EC Number
2.3.1.199
Pubmed
EMBL
BABH01012389
JTDY01000960
KOB75281.1
ODYU01003837
SOQ43090.1
ODYU01008314
+ More
SOQ51794.1 GDQN01006662 JAT84392.1 GDHF01015574 JAI36740.1 GDHF01010979 JAI41335.1 GBXI01008419 GBXI01006261 JAD05873.1 JAD08031.1 JXJN01022417 GAKP01010864 JAC48088.1 GAMC01021294 GAMC01021292 JAB85261.1 JXJN01022416 GAKP01004356 JAC54596.1 GDHF01008780 JAI43534.1 CCAG010007298 CCAG010007299 KQ459586 KPI98123.1 CP012526 ALC45632.1 KK854897 PTY19585.1 AXCM01003970 GEDC01025805 JAS11493.1 KA647151 AFP61780.1 CH477248 EAT46133.1 AAAB01008944 EAA10106.4 OUUW01000005 SPP80764.1 APCN01000639 ATLV01014624 KE524975 KFB39273.1 AXCN02000640 AGBW02011990 OWR45833.1 JXUM01010789 JXUM01010790 JXUM01010791 JXUM01069333 JXUM01069334 JXUM01069335 GAPW01002918 KQ562547 KQ560317 JAC10680.1 KXJ75646.1 KXJ83082.1 CM000070 EAL26992.1 ADMH02002132 ETN58473.1 CH479186 EDW38920.1 CH964272 EDW85037.1 GFDL01000135 JAV34910.1 GBXI01006392 JAD07900.1 AJVK01022764 JRES01001095 KNC25616.1 GAMC01003780 JAC02776.1 KQ414614 KOC68917.1 KZ288222 PBC32085.1 GL762294 EFZ21714.1 SPP80763.1 JRES01000413 KNC31471.1 CH940650 EDW67476.1 GU588088 ADO28044.1 ABLF02039128 GDHC01009670 GDHC01002997 JAQ08959.1 JAQ15632.1 CH933806 EDW14471.1 KRG01025.1 EDW38921.1 EAL26993.1 KK852652 KDR19404.1 HAEC01010038 SBQ78254.1 CH902617 EDV44190.1 KPU80676.1 NEVH01009772 PNF32665.1 GDQN01007992 GDQN01000519 JAT83062.1 JAT90535.1
SOQ51794.1 GDQN01006662 JAT84392.1 GDHF01015574 JAI36740.1 GDHF01010979 JAI41335.1 GBXI01008419 GBXI01006261 JAD05873.1 JAD08031.1 JXJN01022417 GAKP01010864 JAC48088.1 GAMC01021294 GAMC01021292 JAB85261.1 JXJN01022416 GAKP01004356 JAC54596.1 GDHF01008780 JAI43534.1 CCAG010007298 CCAG010007299 KQ459586 KPI98123.1 CP012526 ALC45632.1 KK854897 PTY19585.1 AXCM01003970 GEDC01025805 JAS11493.1 KA647151 AFP61780.1 CH477248 EAT46133.1 AAAB01008944 EAA10106.4 OUUW01000005 SPP80764.1 APCN01000639 ATLV01014624 KE524975 KFB39273.1 AXCN02000640 AGBW02011990 OWR45833.1 JXUM01010789 JXUM01010790 JXUM01010791 JXUM01069333 JXUM01069334 JXUM01069335 GAPW01002918 KQ562547 KQ560317 JAC10680.1 KXJ75646.1 KXJ83082.1 CM000070 EAL26992.1 ADMH02002132 ETN58473.1 CH479186 EDW38920.1 CH964272 EDW85037.1 GFDL01000135 JAV34910.1 GBXI01006392 JAD07900.1 AJVK01022764 JRES01001095 KNC25616.1 GAMC01003780 JAC02776.1 KQ414614 KOC68917.1 KZ288222 PBC32085.1 GL762294 EFZ21714.1 SPP80763.1 JRES01000413 KNC31471.1 CH940650 EDW67476.1 GU588088 ADO28044.1 ABLF02039128 GDHC01009670 GDHC01002997 JAQ08959.1 JAQ15632.1 CH933806 EDW14471.1 KRG01025.1 EDW38921.1 EAL26993.1 KK852652 KDR19404.1 HAEC01010038 SBQ78254.1 CH902617 EDV44190.1 KPU80676.1 NEVH01009772 PNF32665.1 GDQN01007992 GDQN01000519 JAT83062.1 JAT90535.1
Proteomes
UP000005204
UP000037510
UP000092443
UP000092460
UP000078200
UP000092445
+ More
UP000075920 UP000092444 UP000053268 UP000092553 UP000091820 UP000075900 UP000075883 UP000095301 UP000008820 UP000075901 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075885 UP000268350 UP000069272 UP000075840 UP000075881 UP000076408 UP000030765 UP000075886 UP000007151 UP000069940 UP000249989 UP000001819 UP000000673 UP000075880 UP000008744 UP000007798 UP000075884 UP000092462 UP000037069 UP000265140 UP000053825 UP000095300 UP000192223 UP000242457 UP000221080 UP000008792 UP000007819 UP000009192 UP000027135 UP000005203 UP000007801 UP000235965
UP000075920 UP000092444 UP000053268 UP000092553 UP000091820 UP000075900 UP000075883 UP000095301 UP000008820 UP000075901 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075885 UP000268350 UP000069272 UP000075840 UP000075881 UP000076408 UP000030765 UP000075886 UP000007151 UP000069940 UP000249989 UP000001819 UP000000673 UP000075880 UP000008744 UP000007798 UP000075884 UP000092462 UP000037069 UP000265140 UP000053825 UP000095300 UP000192223 UP000242457 UP000221080 UP000008792 UP000007819 UP000009192 UP000027135 UP000005203 UP000007801 UP000235965
PRIDE
Pfam
PF01151 ELO
ProteinModelPortal
H9JQS0
A0A0L7LJB9
A0A2H1VQJ9
A0A2H1WH44
A0A1E1WBM7
A0A0K8VDZ8
+ More
A0A0K8VS01 A0A0A1XAS6 A0A1A9XJ22 A0A1B0BXZ2 A0A034W2T7 W8AWR6 A0A1B0BXZ1 A0A1A9XJ24 A0A034WG82 A0A0K8VXE8 A0A1A9V757 A0A1A9Z493 A0A1A9V750 A0A1A9Z486 A0A182VTN0 A0A1B0FQK1 A0A1B0FQL7 A0A194PYL9 A0A0M4F374 A0A2R7WMF5 A0A1A9W5Z6 A0A182RA82 A0A182M983 A0A1B6CDH2 T1PEI5 Q17HH4 A0A182T880 A0A182X629 A0A182VEV3 A0A182TLQ9 A0A182LJX9 Q7PPN4 A0A182PW57 A0A3B0JEF4 A0A182F0V7 A0A182IB52 A0A182K6I1 A0A182Y8I0 A0A084VMS9 A0A182QAU9 A0A212EWH4 A0A023EP01 Q29BK4 W5J4Z1 A0A182ISK0 B4GP37 B4NJS5 A0A182NB63 A0A1Q3G529 A0A0A1XAY8 A0A1B0D2D9 A0A0L0C049 W8BNS1 A0A3P8ZCE6 A0A0L7RD74 A0A1I8NVN9 A0A1W4WV09 A0A1A9W5Z8 A0A1W4WV14 A0A2A3EJY7 E9ICJ1 A0A3B0JJ32 A0A0L0CGV6 A0A2D0R3G6 B4M165 E3TCK8 J9JX82 A0A1I8M3I9 A0A146M9A3 B4K958 B4GP38 Q29BK5 A0A067R7E0 A0A3P8XC63 A0A1A8H4U3 A0A087ZPK7 B3M000 A0A2J7QVP9 A0A1E1W7T1
A0A0K8VS01 A0A0A1XAS6 A0A1A9XJ22 A0A1B0BXZ2 A0A034W2T7 W8AWR6 A0A1B0BXZ1 A0A1A9XJ24 A0A034WG82 A0A0K8VXE8 A0A1A9V757 A0A1A9Z493 A0A1A9V750 A0A1A9Z486 A0A182VTN0 A0A1B0FQK1 A0A1B0FQL7 A0A194PYL9 A0A0M4F374 A0A2R7WMF5 A0A1A9W5Z6 A0A182RA82 A0A182M983 A0A1B6CDH2 T1PEI5 Q17HH4 A0A182T880 A0A182X629 A0A182VEV3 A0A182TLQ9 A0A182LJX9 Q7PPN4 A0A182PW57 A0A3B0JEF4 A0A182F0V7 A0A182IB52 A0A182K6I1 A0A182Y8I0 A0A084VMS9 A0A182QAU9 A0A212EWH4 A0A023EP01 Q29BK4 W5J4Z1 A0A182ISK0 B4GP37 B4NJS5 A0A182NB63 A0A1Q3G529 A0A0A1XAY8 A0A1B0D2D9 A0A0L0C049 W8BNS1 A0A3P8ZCE6 A0A0L7RD74 A0A1I8NVN9 A0A1W4WV09 A0A1A9W5Z8 A0A1W4WV14 A0A2A3EJY7 E9ICJ1 A0A3B0JJ32 A0A0L0CGV6 A0A2D0R3G6 B4M165 E3TCK8 J9JX82 A0A1I8M3I9 A0A146M9A3 B4K958 B4GP38 Q29BK5 A0A067R7E0 A0A3P8XC63 A0A1A8H4U3 A0A087ZPK7 B3M000 A0A2J7QVP9 A0A1E1W7T1
Ontologies
GO
GO:0102337
GO:0102756
GO:0102336
GO:0006633
GO:0016021
GO:0102338
GO:0009922
GO:0034626
GO:0019367
GO:0030148
GO:0042761
GO:0034625
GO:0030176
GO:0030497
GO:0007110
GO:0051225
GO:0007111
GO:0000915
GO:0007112
GO:0000212
GO:0042811
GO:0005789
GO:0035338
GO:0006636
GO:0006694
GO:0042632
GO:0035209
GO:0005783
GO:0022857
GO:0006508
GO:0005524
GO:0003676
GO:0004813
GO:0006419
GO:0015923
GO:0005634
GO:0006260
PANTHER
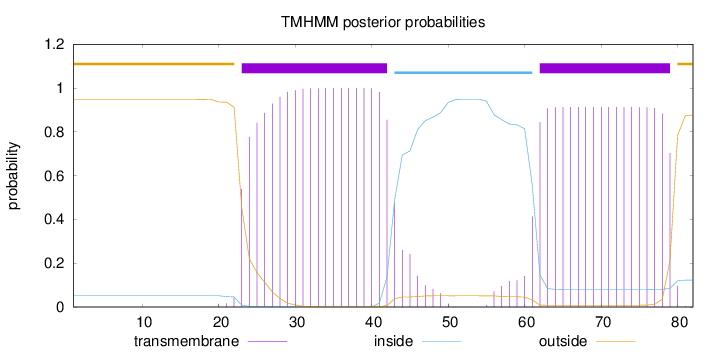
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
82
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
37.35958
Exp number, first 60 AAs:
20.73387
Total prob of N-in:
0.05161
POSSIBLE N-term signal
sequence
outside
1 - 22
TMhelix
23 - 42
inside
43 - 61
TMhelix
62 - 79
outside
80 - 82
Population Genetic Test Statistics
Pi
280.783957
Theta
221.896329
Tajima's D
0.961771
CLR
0
CSRT
0.647467626618669
Interpretation
Uncertain
