Gene
KWMTBOMO06440
Pre Gene Modal
BGIBMGA011877
Annotation
glutamate_[NMDA]_receptor-associated_protein_1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.465
Sequence
CDS
ATGTATCAGAATCAAGGCGGACCGTACCCTCAGGGCGGGGCGTACCCGCAAGGTGGACCGTACCCGCAAGGTGGACCGTACCCACAGGGTGGAGGTTACCCGCAAGGTGGAGCTTACCCTCCAGGAGGAGCGTATCCCCCGGGTGGAGCATACCCGCAGGGCGGATATGCGCCCCCTCAGCCTGGCTTCCAACCTGCATATGGCGGCGCTGGTTACGGTGAACCGGGCTATATCCCACCGAACGACTCAGTCGGCGAAGATGGCGAAGTCAAGGGTTTCGATTTCACTGAAAAATCTATACGCAGGGCTTTCATCCGAAAGGTGTACGCGATCCTGATGTGTCAGCTGATGGTGACGATGGGTTTCATCGCTCTGTTCCTGTACCACCGACCGACCAAAGTCTGGGTGGCCCAAAATCCCTTCATGTTCTGGGTGGCCTTCATAGTGCTGATAGTCTGTCTCATCGCGATGGCGTGCTGTCCCGACGTGAGACGCAAAGCTCCGACGAACTTCATATTCCTGGCCATCTTCACGGCAGCGCAGAGCTTCCTGCTCGGCATTAGCGCAAGCGTCTACGAAGCGGATGCGGTGCTGATGGCAGTTGGCATAACGGCGGCCGTGTGCCTGGGCTTGACTCTGTTCGCTCTGCAGACCAAGTGGGACTTCACCATGATGGGCGGCGTGCTGCTCTGCGCCACCATCGTCCTCCTTGTATTCGGTATCGTGGCCATATTCGTGAAAGGTAAGGTCATAACGCTCGTGTACGCGTCCCTGGGCGCTCTCATATTCTCCATTTACCTCGTCTACGACACCCAGCTGATGATGGGGGGCAAGCACAAGTACAGCATCTCGCCCGAGGAGTACATCTTCGCGGCCCTCAACCTCTACTTGGACATCATCAACATATTCCTGTTCATTCTGACCATCATCGGAGCTTCAAGAGATTAG
Protein
MYQNQGGPYPQGGAYPQGGPYPQGGPYPQGGGYPQGGAYPPGGAYPPGGAYPQGGYAPPQPGFQPAYGGAGYGEPGYIPPNDSVGEDGEVKGFDFTEKSIRRAFIRKVYAILMCQLMVTMGFIALFLYHRPTKVWVAQNPFMFWVAFIVLIVCLIAMACCPDVRRKAPTNFIFLAIFTAAQSFLLGISASVYEADAVLMAVGITAAVCLGLTLFALQTKWDFTMMGGVLLCATIVLLVFGIVAIFVKGKVITLVYASLGALIFSIYLVYDTQLMMGGKHKYSISPEEYIFAALNLYLDIINIFLFILTIIGASRD
Summary
Similarity
Belongs to the BI1 family.
Uniprot
H9JQS1
Q2F693
Q2F692
A0A2A4JM66
A0A2A4JLM1
A0A2W1C3L4
+ More
A0A3S2M196 A0A1L8DK42 A0A1L8DJZ8 A0A1L8DKA9 U5EY08 A0A1Q3EUD2 A0A182SF74 A0A182IQ08 Q5TRL3 A0A182WQT0 A0A1W4XJ16 A0A2C9GQR4 W5JPG5 A0A182TXT8 A0A182X553 A0A182LJH3 A7UTR0 A0A2C9GQR2 A0A2M4BUG9 A0A2M3ZGT0 A0A2M4CRS4 T1DGP6 A0A2M4BTA8 A0A2M3Z2V5 A0A182MVD9 T1E228 A0A182VJ61 T1EAT3 A0A2M4CRY2 A0A023EPM2 Q16QI8 A0A2M4AA33 A0A2M4AA16 A0A1S4FSQ3 A0A1E1W7J0 A0A2M4A946 A0A084VZ81 A0A1E1WGD1 A0A1W4XJF1 A0A1W4XUF6 N6TD50 A0A182NM25 B0X8G1 A0A182RN19 A0A1Q3FMX7 A0A182PP58 J3JY12 U4UT76 A0A182QQZ0 A0A182Y0Z8 A0A182K795 A0A182FHG0 A0A0Q9WVY7 A0A2J7PJT8 B4MJR1 A0A0Q9XJF3 B4KRH2 A0A0C9R6S2 V5G443 Q95T37 Q7JR91 B4HPX7 Q7JRM9 A0A0Q9WF45 A0A0J9U0B9 A0A0J9RBG8 B4LQ63 A0A034WJ36 B4QDI4 A0A0K8WLW1 D3TMS5 A0A1B0BE76 A0A1A9USR0 A0A1A9XN90 A0A034WGH2 A0A1B0A9G2 A0A0K8WDV8 A0A0K8VIN0 A0A0K8URV4 A0A0M4EC74 A0A0A1XD01 A0A1W4VQN1 A0A195E5H6 A0A0Q5VMP3 A0A0A1WV96 A0A0Q5VNB9 A0A1W4VCL5
A0A3S2M196 A0A1L8DK42 A0A1L8DJZ8 A0A1L8DKA9 U5EY08 A0A1Q3EUD2 A0A182SF74 A0A182IQ08 Q5TRL3 A0A182WQT0 A0A1W4XJ16 A0A2C9GQR4 W5JPG5 A0A182TXT8 A0A182X553 A0A182LJH3 A7UTR0 A0A2C9GQR2 A0A2M4BUG9 A0A2M3ZGT0 A0A2M4CRS4 T1DGP6 A0A2M4BTA8 A0A2M3Z2V5 A0A182MVD9 T1E228 A0A182VJ61 T1EAT3 A0A2M4CRY2 A0A023EPM2 Q16QI8 A0A2M4AA33 A0A2M4AA16 A0A1S4FSQ3 A0A1E1W7J0 A0A2M4A946 A0A084VZ81 A0A1E1WGD1 A0A1W4XJF1 A0A1W4XUF6 N6TD50 A0A182NM25 B0X8G1 A0A182RN19 A0A1Q3FMX7 A0A182PP58 J3JY12 U4UT76 A0A182QQZ0 A0A182Y0Z8 A0A182K795 A0A182FHG0 A0A0Q9WVY7 A0A2J7PJT8 B4MJR1 A0A0Q9XJF3 B4KRH2 A0A0C9R6S2 V5G443 Q95T37 Q7JR91 B4HPX7 Q7JRM9 A0A0Q9WF45 A0A0J9U0B9 A0A0J9RBG8 B4LQ63 A0A034WJ36 B4QDI4 A0A0K8WLW1 D3TMS5 A0A1B0BE76 A0A1A9USR0 A0A1A9XN90 A0A034WGH2 A0A1B0A9G2 A0A0K8WDV8 A0A0K8VIN0 A0A0K8URV4 A0A0M4EC74 A0A0A1XD01 A0A1W4VQN1 A0A195E5H6 A0A0Q5VMP3 A0A0A1WV96 A0A0Q5VNB9 A0A1W4VCL5
Pubmed
EMBL
BABH01012391
DQ311179
ABD36124.1
DQ311180
ABD36125.1
NWSH01001090
+ More
PCG72674.1 PCG72676.1 KZ149896 PZC78753.1 RSAL01000076 RVE48854.1 GFDF01007303 JAV06781.1 GFDF01007302 JAV06782.1 GFDF01007304 JAV06780.1 GANO01000662 JAB59209.1 GFDL01016124 JAV18921.1 AAAB01008960 EAL39852.3 APCN01004290 ADMH02000483 ETN66287.1 EDO63682.1 EDO63683.1 GGFJ01007501 MBW56642.1 GGFM01006897 MBW27648.1 GGFL01003862 MBW68040.1 GALA01001581 JAA93271.1 GGFJ01007093 MBW56234.1 GGFM01002096 MBW22847.1 AXCM01003248 GALA01001580 JAA93272.1 GAMD01000768 JAB00823.1 GGFL01003861 MBW68039.1 GAPW01002625 JAC10973.1 CH477747 EAT36652.1 GGFK01004281 MBW37602.1 GGFK01004280 MBW37601.1 GDQN01008140 JAT82914.1 GGFK01003929 MBW37250.1 ATLV01018617 KE525239 KFB43275.1 GDQN01011336 GDQN01005001 JAT79718.1 JAT86053.1 APGK01042700 KB741007 ENN75623.1 DS232488 EDS42497.1 GFDL01006094 JAV28951.1 BT128135 GABX01000002 AEE63096.1 JAA74517.1 KB631811 KI207771 ERL86343.1 ERL95733.1 AXCN02002327 CH963846 KRF97540.1 NEVH01024946 PNF16588.1 EDW72350.1 CH933808 KRG05218.1 EDW10398.1 GBYB01012034 JAG81801.1 GALX01003661 JAB64805.1 AY060346 AE013599 AAL25385.1 AAM68615.1 AGB93462.1 BT003175 BT015290 AAM68612.1 AAM68613.1 AAO24930.1 AAT94519.1 AFH08042.1 AFH08043.1 CH480816 EDW47640.1 BT001413 AAF58446.1 AAM68614.1 AAM68616.2 AAN71168.1 CH940648 KRF79935.1 CM002911 KMY93365.1 KMY93363.1 KMY93364.1 KMY93366.1 EDW61348.1 GAKP01005169 JAC53783.1 CM000362 EDX06849.1 KMY93361.1 GDHF01000469 JAI51845.1 EZ422727 ADD19003.1 JXJN01012807 GAKP01005168 JAC53784.1 GDHF01003047 JAI49267.1 GDHF01022575 GDHF01013601 GDHF01012866 JAI29739.1 JAI38713.1 JAI39448.1 GDHF01022880 GDHF01019505 GDHF01003837 JAI29434.1 JAI32809.1 JAI48477.1 CP012524 ALC41277.1 GBXI01005486 JAD08806.1 KQ979609 KYN20152.1 CH954179 KQS62780.1 KQS62782.1 KQS62784.1 GBXI01011989 JAD02303.1 KQS62785.1
PCG72674.1 PCG72676.1 KZ149896 PZC78753.1 RSAL01000076 RVE48854.1 GFDF01007303 JAV06781.1 GFDF01007302 JAV06782.1 GFDF01007304 JAV06780.1 GANO01000662 JAB59209.1 GFDL01016124 JAV18921.1 AAAB01008960 EAL39852.3 APCN01004290 ADMH02000483 ETN66287.1 EDO63682.1 EDO63683.1 GGFJ01007501 MBW56642.1 GGFM01006897 MBW27648.1 GGFL01003862 MBW68040.1 GALA01001581 JAA93271.1 GGFJ01007093 MBW56234.1 GGFM01002096 MBW22847.1 AXCM01003248 GALA01001580 JAA93272.1 GAMD01000768 JAB00823.1 GGFL01003861 MBW68039.1 GAPW01002625 JAC10973.1 CH477747 EAT36652.1 GGFK01004281 MBW37602.1 GGFK01004280 MBW37601.1 GDQN01008140 JAT82914.1 GGFK01003929 MBW37250.1 ATLV01018617 KE525239 KFB43275.1 GDQN01011336 GDQN01005001 JAT79718.1 JAT86053.1 APGK01042700 KB741007 ENN75623.1 DS232488 EDS42497.1 GFDL01006094 JAV28951.1 BT128135 GABX01000002 AEE63096.1 JAA74517.1 KB631811 KI207771 ERL86343.1 ERL95733.1 AXCN02002327 CH963846 KRF97540.1 NEVH01024946 PNF16588.1 EDW72350.1 CH933808 KRG05218.1 EDW10398.1 GBYB01012034 JAG81801.1 GALX01003661 JAB64805.1 AY060346 AE013599 AAL25385.1 AAM68615.1 AGB93462.1 BT003175 BT015290 AAM68612.1 AAM68613.1 AAO24930.1 AAT94519.1 AFH08042.1 AFH08043.1 CH480816 EDW47640.1 BT001413 AAF58446.1 AAM68614.1 AAM68616.2 AAN71168.1 CH940648 KRF79935.1 CM002911 KMY93365.1 KMY93363.1 KMY93364.1 KMY93366.1 EDW61348.1 GAKP01005169 JAC53783.1 CM000362 EDX06849.1 KMY93361.1 GDHF01000469 JAI51845.1 EZ422727 ADD19003.1 JXJN01012807 GAKP01005168 JAC53784.1 GDHF01003047 JAI49267.1 GDHF01022575 GDHF01013601 GDHF01012866 JAI29739.1 JAI38713.1 JAI39448.1 GDHF01022880 GDHF01019505 GDHF01003837 JAI29434.1 JAI32809.1 JAI48477.1 CP012524 ALC41277.1 GBXI01005486 JAD08806.1 KQ979609 KYN20152.1 CH954179 KQS62780.1 KQS62782.1 KQS62784.1 GBXI01011989 JAD02303.1 KQS62785.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000075901
UP000075880
UP000007062
+ More
UP000075920 UP000192223 UP000075840 UP000000673 UP000075902 UP000076407 UP000075882 UP000075883 UP000075903 UP000008820 UP000030765 UP000019118 UP000075884 UP000002320 UP000075900 UP000075885 UP000030742 UP000075886 UP000076408 UP000075881 UP000069272 UP000007798 UP000235965 UP000009192 UP000000803 UP000001292 UP000008792 UP000000304 UP000092460 UP000078200 UP000092443 UP000092445 UP000092553 UP000192221 UP000078492 UP000008711
UP000075920 UP000192223 UP000075840 UP000000673 UP000075902 UP000076407 UP000075882 UP000075883 UP000075903 UP000008820 UP000030765 UP000019118 UP000075884 UP000002320 UP000075900 UP000075885 UP000030742 UP000075886 UP000076408 UP000075881 UP000069272 UP000007798 UP000235965 UP000009192 UP000000803 UP000001292 UP000008792 UP000000304 UP000092460 UP000078200 UP000092443 UP000092445 UP000092553 UP000192221 UP000078492 UP000008711
Pfam
PF01027 Bax1-I
Interpro
IPR006214
Bax_inhibitor_1-related
ProteinModelPortal
H9JQS1
Q2F693
Q2F692
A0A2A4JM66
A0A2A4JLM1
A0A2W1C3L4
+ More
A0A3S2M196 A0A1L8DK42 A0A1L8DJZ8 A0A1L8DKA9 U5EY08 A0A1Q3EUD2 A0A182SF74 A0A182IQ08 Q5TRL3 A0A182WQT0 A0A1W4XJ16 A0A2C9GQR4 W5JPG5 A0A182TXT8 A0A182X553 A0A182LJH3 A7UTR0 A0A2C9GQR2 A0A2M4BUG9 A0A2M3ZGT0 A0A2M4CRS4 T1DGP6 A0A2M4BTA8 A0A2M3Z2V5 A0A182MVD9 T1E228 A0A182VJ61 T1EAT3 A0A2M4CRY2 A0A023EPM2 Q16QI8 A0A2M4AA33 A0A2M4AA16 A0A1S4FSQ3 A0A1E1W7J0 A0A2M4A946 A0A084VZ81 A0A1E1WGD1 A0A1W4XJF1 A0A1W4XUF6 N6TD50 A0A182NM25 B0X8G1 A0A182RN19 A0A1Q3FMX7 A0A182PP58 J3JY12 U4UT76 A0A182QQZ0 A0A182Y0Z8 A0A182K795 A0A182FHG0 A0A0Q9WVY7 A0A2J7PJT8 B4MJR1 A0A0Q9XJF3 B4KRH2 A0A0C9R6S2 V5G443 Q95T37 Q7JR91 B4HPX7 Q7JRM9 A0A0Q9WF45 A0A0J9U0B9 A0A0J9RBG8 B4LQ63 A0A034WJ36 B4QDI4 A0A0K8WLW1 D3TMS5 A0A1B0BE76 A0A1A9USR0 A0A1A9XN90 A0A034WGH2 A0A1B0A9G2 A0A0K8WDV8 A0A0K8VIN0 A0A0K8URV4 A0A0M4EC74 A0A0A1XD01 A0A1W4VQN1 A0A195E5H6 A0A0Q5VMP3 A0A0A1WV96 A0A0Q5VNB9 A0A1W4VCL5
A0A3S2M196 A0A1L8DK42 A0A1L8DJZ8 A0A1L8DKA9 U5EY08 A0A1Q3EUD2 A0A182SF74 A0A182IQ08 Q5TRL3 A0A182WQT0 A0A1W4XJ16 A0A2C9GQR4 W5JPG5 A0A182TXT8 A0A182X553 A0A182LJH3 A7UTR0 A0A2C9GQR2 A0A2M4BUG9 A0A2M3ZGT0 A0A2M4CRS4 T1DGP6 A0A2M4BTA8 A0A2M3Z2V5 A0A182MVD9 T1E228 A0A182VJ61 T1EAT3 A0A2M4CRY2 A0A023EPM2 Q16QI8 A0A2M4AA33 A0A2M4AA16 A0A1S4FSQ3 A0A1E1W7J0 A0A2M4A946 A0A084VZ81 A0A1E1WGD1 A0A1W4XJF1 A0A1W4XUF6 N6TD50 A0A182NM25 B0X8G1 A0A182RN19 A0A1Q3FMX7 A0A182PP58 J3JY12 U4UT76 A0A182QQZ0 A0A182Y0Z8 A0A182K795 A0A182FHG0 A0A0Q9WVY7 A0A2J7PJT8 B4MJR1 A0A0Q9XJF3 B4KRH2 A0A0C9R6S2 V5G443 Q95T37 Q7JR91 B4HPX7 Q7JRM9 A0A0Q9WF45 A0A0J9U0B9 A0A0J9RBG8 B4LQ63 A0A034WJ36 B4QDI4 A0A0K8WLW1 D3TMS5 A0A1B0BE76 A0A1A9USR0 A0A1A9XN90 A0A034WGH2 A0A1B0A9G2 A0A0K8WDV8 A0A0K8VIN0 A0A0K8URV4 A0A0M4EC74 A0A0A1XD01 A0A1W4VQN1 A0A195E5H6 A0A0Q5VMP3 A0A0A1WV96 A0A0Q5VNB9 A0A1W4VCL5
Ontologies
PANTHER
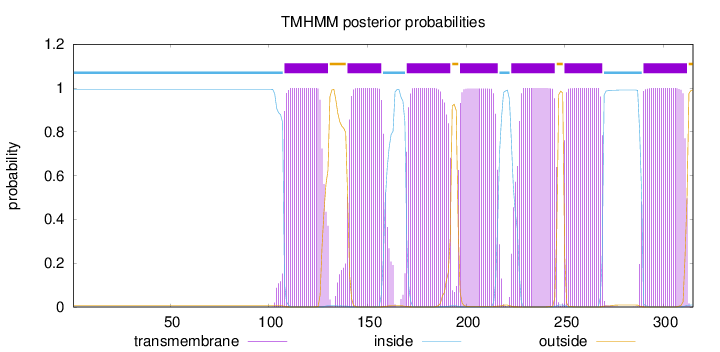
Topology
Length:
315
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
148.46318
Exp number, first 60 AAs:
0.00127
Total prob of N-in:
0.99404
inside
1 - 107
TMhelix
108 - 130
outside
131 - 139
TMhelix
140 - 157
inside
158 - 169
TMhelix
170 - 192
outside
193 - 196
TMhelix
197 - 216
inside
217 - 222
TMhelix
223 - 245
outside
246 - 249
TMhelix
250 - 269
inside
270 - 289
TMhelix
290 - 312
outside
313 - 315
Population Genetic Test Statistics
Pi
215.269076
Theta
166.150117
Tajima's D
1.084056
CLR
0.665264
CSRT
0.674116294185291
Interpretation
Uncertain
