Gene
KWMTBOMO06436
Pre Gene Modal
BGIBMGA011605
Annotation
PREDICTED:_dyslexia_susceptibility_1_candidate_gene_1_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.799
Sequence
CDS
ATGCCGATAATTGTTAAAGATTTTACTTGGACGCAAACTGAAACTACAGTTCATATTAGGATCCCATTAAATCCGGGGAAACATGATAAAGTAGATTTGTTTACCACGGACTCTTACATAAAAGCGCATTTTAAACCATTTCTTTTCGAAGTCTTCCTACGATACGATGTTAACATAAGCAAAAGTAAATGTATTATTAATGACGACGAAATTACTTTGGATTTGTTAAAAAACAATGAAGAACAGTGGGATGGTCTAGAAAAGAATTTGAGTAAAGAAGAAAAAAAGGTTCTAAGGGATGAAGTTTATAAAAAGAGCCAAGAAAGGGCAAAAGAAGATGCGGAAAACAGATCTATTAAAAAAAGTCAATTAGACAGGTTTACAGTACAAAGAGCTATGGATTTAGATACAAAACAACATGCTCTGATGGATTCTAGAAGAGATCACGAGAGATATCAAGCTATGAATGCTTTGGAAGAGTGGAGACAACAGACAAACAAGAATGATTATGATGTAAATGAAAGCGGTGTTAAAATTACCGAAATGTCAGAGAATGAGCCTGTAAAAGTAAAAATAAGTACAGAAAAAGCACAATCAAAAGTGCGACCCCAAGCTACAACCCCAAGAACACCTCTGAAGACACCTGTGAAGTCGGAATTTGTAGAAAAAAAGAAGAAAGAAGCAGCAACTAGAGTTTTACCGAAATTGAGGCAAACCGCGCACTTGGAAATAACTCACACACCCAGATCATTTCCAACACCGAGCAGAGAATCCACAGCAGAGGAAGAGCAGACTTGGTTGAAGAACATCACCATGGCTCGTAGAGCTACGGGTTTCGTATCGGAAGACTTAAGGCCAGAGGAACAGGACCCTCAGTGGTGCAAAGAGAAGGGAGACGAATTCTTCCGCAACGGTAACTTTTTGGGAGCAATCAGTGCGTACACTCATGGCATAAAGTTGTCCGCGAAGTTGCCCAGTTTGTATTCGAACAGGGCTGCAACGCATTTCGCCCTCGGAAATTTTAATAAATGCGTAAACGACTGCTCCACAGCGTTAGACCTCCTGAAGCCGGCGTGTGAAGGGAACCGTAAGAGTCGAGCTAGATGCATCGCGAGAAGGGCCGCGGCTCTGGCCAGGCTCGGCTATCTGAACAAAGCCATTGAGGAAATGAAGGCGGCCAGTAGATTGATACCCGATGACCAAAAAATTAAGAAGGATATTTACGATATGGAAAGAGCTTGGGAACAAAATCCGGATTCCGATTGA
Protein
MPIIVKDFTWTQTETTVHIRIPLNPGKHDKVDLFTTDSYIKAHFKPFLFEVFLRYDVNISKSKCIINDDEITLDLLKNNEEQWDGLEKNLSKEEKKVLRDEVYKKSQERAKEDAENRSIKKSQLDRFTVQRAMDLDTKQHALMDSRRDHERYQAMNALEEWRQQTNKNDYDVNESGVKITEMSENEPVKVKISTEKAQSKVRPQATTPRTPLKTPVKSEFVEKKKKEAATRVLPKLRQTAHLEITHTPRSFPTPSRESTAEEEQTWLKNITMARRATGFVSEDLRPEEQDPQWCKEKGDEFFRNGNFLGAISAYTHGIKLSAKLPSLYSNRAATHFALGNFNKCVNDCSTALDLLKPACEGNRKSRARCIARRAAALARLGYLNKAIEEMKAASRLIPDDQKIKKDIYDMERAWEQNPDSD
Summary
Similarity
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Uniprot
H9JQ00
A0A2A4JKU1
A0A2H1W3K7
A0A2W1BUV2
A0A212FK71
A0A0N1I5N6
+ More
A0A194PV34 A0A0L7LP28 A0A067QW13 A0A2J7QGW7 E0VVV3 A0A2J7QGW3 A0A2J7QGV7 D6WGD9 A0A1W4WMZ4 A0A088ARE7 J9K2Q1 A0A1B6KSX5 Q17PU2 A0A3B4D256 A0A1S3KVU8 A0A0R4ID80 U4UIY8 Q6P010 A0A1S3J8Z5 A0A3Q1HQX2 A0A182WSU2 Q4JS98 Q5VJR9 A0A3Q2E173 A0A3Q2FS64 N6TTB8 A0A0A9ZCE5 A0A182HLP5 A0A182T455 A0A182NRT0 A0A3N0XRH3 A0A146LYM9 A0A060XHA0 A0A182U9U0 A0A182RZJ9 A0A3Q3ETH5 A0A1L8H0M0 A0A182FPW1 Q5VJS2 A0A182VZM9 Q6AZN0 A0A3B3QDT1 V9KW27 A0A182JU76 W5UF71 K1PPK0 A0A2U9BZ66 A0A2J7QGW1 A0A3B1JIU1 A0A3S2PCT8 A0A182JL43 A0A3P8UEY9 A0A3Q3M7B4 A0A182V3L3 A0A182M6D7 A0A2I4CEL2 A0A336M803 A0A3Q3LSA0 A0A0B6ZTP8 A0A3Q1FNM6 W5N7K3 A0A1W4YUC3 A0A3Q2V976 A0A182LA39 A0A3S1A0H3 A0A3B3W076 Q5VJS3 A0A182GC27 A0A3Q2YED5 V4AZK8 A0A3P8Y624 A0A3P8QCM7 A0A1S3DD38 A0A3P9D7V5 A0A3P9Q5V5 A0A3Q1CM12 H2MHW7 I3K4E2 A0A3P8S1Z5 A0A3Q4GNL6 A0A3B4FHW3 A0A0P7XLH6 A0A3B5M1Z7 A0A3B3XZ68 H2YFR5 I3K4E1 A0A3B4FHX4 A0A3Q7S7Y6 A0A1B6I079 A0A3B4TBJ7 A0A087Y9P2 G3PM27 A0A315WX37 A0A3B4WSQ9
A0A194PV34 A0A0L7LP28 A0A067QW13 A0A2J7QGW7 E0VVV3 A0A2J7QGW3 A0A2J7QGV7 D6WGD9 A0A1W4WMZ4 A0A088ARE7 J9K2Q1 A0A1B6KSX5 Q17PU2 A0A3B4D256 A0A1S3KVU8 A0A0R4ID80 U4UIY8 Q6P010 A0A1S3J8Z5 A0A3Q1HQX2 A0A182WSU2 Q4JS98 Q5VJR9 A0A3Q2E173 A0A3Q2FS64 N6TTB8 A0A0A9ZCE5 A0A182HLP5 A0A182T455 A0A182NRT0 A0A3N0XRH3 A0A146LYM9 A0A060XHA0 A0A182U9U0 A0A182RZJ9 A0A3Q3ETH5 A0A1L8H0M0 A0A182FPW1 Q5VJS2 A0A182VZM9 Q6AZN0 A0A3B3QDT1 V9KW27 A0A182JU76 W5UF71 K1PPK0 A0A2U9BZ66 A0A2J7QGW1 A0A3B1JIU1 A0A3S2PCT8 A0A182JL43 A0A3P8UEY9 A0A3Q3M7B4 A0A182V3L3 A0A182M6D7 A0A2I4CEL2 A0A336M803 A0A3Q3LSA0 A0A0B6ZTP8 A0A3Q1FNM6 W5N7K3 A0A1W4YUC3 A0A3Q2V976 A0A182LA39 A0A3S1A0H3 A0A3B3W076 Q5VJS3 A0A182GC27 A0A3Q2YED5 V4AZK8 A0A3P8Y624 A0A3P8QCM7 A0A1S3DD38 A0A3P9D7V5 A0A3P9Q5V5 A0A3Q1CM12 H2MHW7 I3K4E2 A0A3P8S1Z5 A0A3Q4GNL6 A0A3B4FHW3 A0A0P7XLH6 A0A3B5M1Z7 A0A3B3XZ68 H2YFR5 I3K4E1 A0A3B4FHX4 A0A3Q7S7Y6 A0A1B6I079 A0A3B4TBJ7 A0A087Y9P2 G3PM27 A0A315WX37 A0A3B4WSQ9
Pubmed
19121390
28756777
22118469
26354079
26227816
24845553
+ More
20566863 18362917 19820115 17510324 23594743 23537049 15944077 12364791 14747013 17210077 25401762 26823975 24755649 27762356 29240929 24402279 23127152 22992520 25329095 24487278 20966253 20431018 26483478 23254933 25069045 25186727 17554307 29703783
20566863 18362917 19820115 17510324 23594743 23537049 15944077 12364791 14747013 17210077 25401762 26823975 24755649 27762356 29240929 24402279 23127152 22992520 25329095 24487278 20966253 20431018 26483478 23254933 25069045 25186727 17554307 29703783
EMBL
BABH01012392
NWSH01001090
PCG72667.1
ODYU01006045
SOQ47546.1
KZ149896
+ More
PZC78749.1 AGBW02008141 OWR54122.1 KQ460971 KPJ10130.1 KQ459590 KPI97182.1 JTDY01000437 KOB77170.1 KK852879 KDR14473.1 NEVH01014359 PNF27820.1 DS235815 EEB17509.1 PNF27825.1 PNF27824.1 KQ971329 EFA01154.1 ABLF02005734 ABLF02005736 ABLF02005737 ABLF02005738 ABLF02028634 ABLF02028643 ABLF02063984 GEBQ01025476 JAT14501.1 CH477189 EAT48749.1 FO704795 KB632344 ERL93042.1 BC065881 BC165842 AY428510 AAH65881.1 AAI65842.1 AAR89528.1 CR954257 CAJ14164.1 AY428512 AAAB01008987 AAR89530.1 EAU75637.1 APGK01055167 KB741261 ENN71581.1 GBHO01002586 JAG41018.1 APCN01000909 RJVU01062584 ROJ29156.1 GDHC01006837 JAQ11792.1 FR905394 CDQ78998.1 CM004470 OCT89561.1 AY428509 AAR89527.1 BC077575 AAH77575.1 JW870735 AFP03253.1 JT414460 AHH40628.1 JH819193 EKC23538.1 CP026252 AWP08990.1 PNF27821.1 CM012442 RVE72169.1 AXCM01000025 AXCM01000026 UFQT01000524 SSX24973.1 HACG01024361 CEK71226.1 AHAT01024879 RQTK01000443 RUS79575.1 AAMC01125559 AAMC01125560 BC170925 BC170927 AY428508 AAI70925.1 AAR89526.1 JXUM01053620 JXUM01053621 JXUM01053622 KQ561784 KXJ77550.1 KB200701 ESP00556.1 AERX01031820 AERX01031821 JARO02001277 KPP75954.1 GECU01027378 JAS80328.1 AYCK01011943 NHOQ01000086 PWA33134.1
PZC78749.1 AGBW02008141 OWR54122.1 KQ460971 KPJ10130.1 KQ459590 KPI97182.1 JTDY01000437 KOB77170.1 KK852879 KDR14473.1 NEVH01014359 PNF27820.1 DS235815 EEB17509.1 PNF27825.1 PNF27824.1 KQ971329 EFA01154.1 ABLF02005734 ABLF02005736 ABLF02005737 ABLF02005738 ABLF02028634 ABLF02028643 ABLF02063984 GEBQ01025476 JAT14501.1 CH477189 EAT48749.1 FO704795 KB632344 ERL93042.1 BC065881 BC165842 AY428510 AAH65881.1 AAI65842.1 AAR89528.1 CR954257 CAJ14164.1 AY428512 AAAB01008987 AAR89530.1 EAU75637.1 APGK01055167 KB741261 ENN71581.1 GBHO01002586 JAG41018.1 APCN01000909 RJVU01062584 ROJ29156.1 GDHC01006837 JAQ11792.1 FR905394 CDQ78998.1 CM004470 OCT89561.1 AY428509 AAR89527.1 BC077575 AAH77575.1 JW870735 AFP03253.1 JT414460 AHH40628.1 JH819193 EKC23538.1 CP026252 AWP08990.1 PNF27821.1 CM012442 RVE72169.1 AXCM01000025 AXCM01000026 UFQT01000524 SSX24973.1 HACG01024361 CEK71226.1 AHAT01024879 RQTK01000443 RUS79575.1 AAMC01125559 AAMC01125560 BC170925 BC170927 AY428508 AAI70925.1 AAR89526.1 JXUM01053620 JXUM01053621 JXUM01053622 KQ561784 KXJ77550.1 KB200701 ESP00556.1 AERX01031820 AERX01031821 JARO02001277 KPP75954.1 GECU01027378 JAS80328.1 AYCK01011943 NHOQ01000086 PWA33134.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000027135 UP000235965 UP000009046 UP000007266 UP000192223 UP000005203 UP000007819 UP000008820 UP000261440 UP000087266 UP000000437 UP000030742 UP000085678 UP000265040 UP000076407 UP000007062 UP000265020 UP000019118 UP000075840 UP000075901 UP000075884 UP000193380 UP000075902 UP000075900 UP000261660 UP000186698 UP000069272 UP000075920 UP000261540 UP000075881 UP000221080 UP000005408 UP000246464 UP000018467 UP000075880 UP000265120 UP000261640 UP000075903 UP000075883 UP000192220 UP000257200 UP000018468 UP000192224 UP000264840 UP000075882 UP000271974 UP000261500 UP000008143 UP000069940 UP000249989 UP000264820 UP000030746 UP000265140 UP000265100 UP000079169 UP000265160 UP000242638 UP000257160 UP000001038 UP000005207 UP000265080 UP000261580 UP000261460 UP000034805 UP000261380 UP000261480 UP000007875 UP000286640 UP000261420 UP000028760 UP000007635 UP000261360
UP000027135 UP000235965 UP000009046 UP000007266 UP000192223 UP000005203 UP000007819 UP000008820 UP000261440 UP000087266 UP000000437 UP000030742 UP000085678 UP000265040 UP000076407 UP000007062 UP000265020 UP000019118 UP000075840 UP000075901 UP000075884 UP000193380 UP000075902 UP000075900 UP000261660 UP000186698 UP000069272 UP000075920 UP000261540 UP000075881 UP000221080 UP000005408 UP000246464 UP000018467 UP000075880 UP000265120 UP000261640 UP000075903 UP000075883 UP000192220 UP000257200 UP000018468 UP000192224 UP000264840 UP000075882 UP000271974 UP000261500 UP000008143 UP000069940 UP000249989 UP000264820 UP000030746 UP000265140 UP000265100 UP000079169 UP000265160 UP000242638 UP000257160 UP000001038 UP000005207 UP000265080 UP000261580 UP000261460 UP000034805 UP000261380 UP000261480 UP000007875 UP000286640 UP000261420 UP000028760 UP000007635 UP000261360
PRIDE
Interpro
IPR011990
TPR-like_helical_dom_sf
+ More
IPR007052 CS_dom
IPR019734 TPR_repeat
IPR008978 HSP20-like_chaperone
IPR013026 TPR-contain_dom
IPR037894 CS_DYX1C1
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR013105 TPR_2
IPR023395 Mt_carrier_dom_sf
IPR018108 Mitochondrial_sb/sol_carrier
IPR001440 TPR_1
IPR007052 CS_dom
IPR019734 TPR_repeat
IPR008978 HSP20-like_chaperone
IPR013026 TPR-contain_dom
IPR037894 CS_DYX1C1
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR013105 TPR_2
IPR023395 Mt_carrier_dom_sf
IPR018108 Mitochondrial_sb/sol_carrier
IPR001440 TPR_1
Gene 3D
ProteinModelPortal
H9JQ00
A0A2A4JKU1
A0A2H1W3K7
A0A2W1BUV2
A0A212FK71
A0A0N1I5N6
+ More
A0A194PV34 A0A0L7LP28 A0A067QW13 A0A2J7QGW7 E0VVV3 A0A2J7QGW3 A0A2J7QGV7 D6WGD9 A0A1W4WMZ4 A0A088ARE7 J9K2Q1 A0A1B6KSX5 Q17PU2 A0A3B4D256 A0A1S3KVU8 A0A0R4ID80 U4UIY8 Q6P010 A0A1S3J8Z5 A0A3Q1HQX2 A0A182WSU2 Q4JS98 Q5VJR9 A0A3Q2E173 A0A3Q2FS64 N6TTB8 A0A0A9ZCE5 A0A182HLP5 A0A182T455 A0A182NRT0 A0A3N0XRH3 A0A146LYM9 A0A060XHA0 A0A182U9U0 A0A182RZJ9 A0A3Q3ETH5 A0A1L8H0M0 A0A182FPW1 Q5VJS2 A0A182VZM9 Q6AZN0 A0A3B3QDT1 V9KW27 A0A182JU76 W5UF71 K1PPK0 A0A2U9BZ66 A0A2J7QGW1 A0A3B1JIU1 A0A3S2PCT8 A0A182JL43 A0A3P8UEY9 A0A3Q3M7B4 A0A182V3L3 A0A182M6D7 A0A2I4CEL2 A0A336M803 A0A3Q3LSA0 A0A0B6ZTP8 A0A3Q1FNM6 W5N7K3 A0A1W4YUC3 A0A3Q2V976 A0A182LA39 A0A3S1A0H3 A0A3B3W076 Q5VJS3 A0A182GC27 A0A3Q2YED5 V4AZK8 A0A3P8Y624 A0A3P8QCM7 A0A1S3DD38 A0A3P9D7V5 A0A3P9Q5V5 A0A3Q1CM12 H2MHW7 I3K4E2 A0A3P8S1Z5 A0A3Q4GNL6 A0A3B4FHW3 A0A0P7XLH6 A0A3B5M1Z7 A0A3B3XZ68 H2YFR5 I3K4E1 A0A3B4FHX4 A0A3Q7S7Y6 A0A1B6I079 A0A3B4TBJ7 A0A087Y9P2 G3PM27 A0A315WX37 A0A3B4WSQ9
A0A194PV34 A0A0L7LP28 A0A067QW13 A0A2J7QGW7 E0VVV3 A0A2J7QGW3 A0A2J7QGV7 D6WGD9 A0A1W4WMZ4 A0A088ARE7 J9K2Q1 A0A1B6KSX5 Q17PU2 A0A3B4D256 A0A1S3KVU8 A0A0R4ID80 U4UIY8 Q6P010 A0A1S3J8Z5 A0A3Q1HQX2 A0A182WSU2 Q4JS98 Q5VJR9 A0A3Q2E173 A0A3Q2FS64 N6TTB8 A0A0A9ZCE5 A0A182HLP5 A0A182T455 A0A182NRT0 A0A3N0XRH3 A0A146LYM9 A0A060XHA0 A0A182U9U0 A0A182RZJ9 A0A3Q3ETH5 A0A1L8H0M0 A0A182FPW1 Q5VJS2 A0A182VZM9 Q6AZN0 A0A3B3QDT1 V9KW27 A0A182JU76 W5UF71 K1PPK0 A0A2U9BZ66 A0A2J7QGW1 A0A3B1JIU1 A0A3S2PCT8 A0A182JL43 A0A3P8UEY9 A0A3Q3M7B4 A0A182V3L3 A0A182M6D7 A0A2I4CEL2 A0A336M803 A0A3Q3LSA0 A0A0B6ZTP8 A0A3Q1FNM6 W5N7K3 A0A1W4YUC3 A0A3Q2V976 A0A182LA39 A0A3S1A0H3 A0A3B3W076 Q5VJS3 A0A182GC27 A0A3Q2YED5 V4AZK8 A0A3P8Y624 A0A3P8QCM7 A0A1S3DD38 A0A3P9D7V5 A0A3P9Q5V5 A0A3Q1CM12 H2MHW7 I3K4E2 A0A3P8S1Z5 A0A3Q4GNL6 A0A3B4FHW3 A0A0P7XLH6 A0A3B5M1Z7 A0A3B3XZ68 H2YFR5 I3K4E1 A0A3B4FHX4 A0A3Q7S7Y6 A0A1B6I079 A0A3B4TBJ7 A0A087Y9P2 G3PM27 A0A315WX37 A0A3B4WSQ9
PDB
2LNI
E-value=0.000261702,
Score=105
Ontologies
GO
Topology
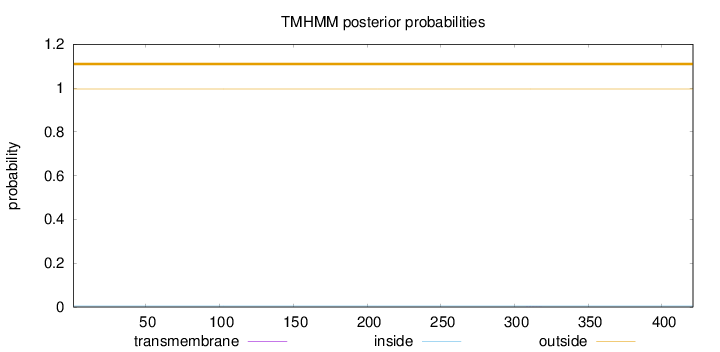
Length:
421
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00282
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00407
outside
1 - 421
Population Genetic Test Statistics
Pi
30.532958
Theta
24.96846
Tajima's D
0.221936
CLR
10.81058
CSRT
0.43602819859007
Interpretation
Uncertain
