Gene
KWMTBOMO06435 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011880
Annotation
phosphotriesterase-like_protein_[Bombyx_mori]
Full name
Phosphotriesterase-related protein
Alternative Name
Parathion hydrolase-related protein
Location in the cell
Cytoplasmic Reliability : 3.067
Sequence
CDS
ATGACTTCAAAAGTAGAAACTGTGTTGGGGCCGGTGCCGATAAGTGTCTTGGGTCGAACTTTAACCCACGAGCATCTAGCGATGACCTTCACCCACTTCTACAGAGAGCCACCAAAGCAAATTGTCGATGTATTCGAGAACCCCTTTTGTCTGGAAAAATCTGGATTTCTAAGACAATTCCCTTACAGTTCTAAGGGTAATTTGGTATTGAATGATCAGGCCGCGAAGGAGGCAGTTGTAAATGATGTTATAGCGTACAAGAAGTTCGGTGGAGGTACAATAGTGGAAAATTCGACGGTCGGTCTTGATCGGGATGTAAACTTTTACAAGTTGGTATCGGAAAAGAGCGACGTAAACATAGTCGCGGGCACCGGCTTCTATGTCGAGGACGTTCAAGCTAAGGCCAACCTTAGCAAAACAACGGAACAAATGTATCATCACATGCTGACTGAACTCACAACAGGGTTTCTTGAGGAACCTAGTGTGAAGGCTGGGTTCATGGGAGAAATAGCCAGTGTTTGGCCATTAAGAGAGTTTGAACGTAGAGCCATCGTAGCTGCCGGTGAGCTGCAGCCGCAAGTCGGCTGCGGTGTCAGTTTCCATCCTCACCGCGCCATCGAAGCTCCATTTGAAATCATTCGTCTGTACCTTGAGGCCGGAGGAAGCAAGGATAAAGTTGTCATGTCGCATTTAGACCGAACCCTACTTGAAGACGACAAATTACTAGAATTCTCTGAGCTCGGTACTTACTGCCAGTTCGATCTGTTTGGCGTGGAAGTCTCATTCTACCAGTTAAACCCGGCGGCTGACATGCCATCAGACGGGCAGCGGATTGATAAAATCAAATTGTTGATCGACGATGGGAAATCTGATAAAGTGTTAATGTCCCACGACATTCACACTAAACATAGATTGACGGCATTCGGTGGCCACGGCTACGCTCACATCATAAACACAGTGCTGCCGAAAATGGCGCTCAAAGGAATATCTCAAGATACCATAGACAGGATTACTATAGACAATCCAGCAAGATGGTTGGCCACTAACATTTAG
Protein
MTSKVETVLGPVPISVLGRTLTHEHLAMTFTHFYREPPKQIVDVFENPFCLEKSGFLRQFPYSSKGNLVLNDQAAKEAVVNDVIAYKKFGGGTIVENSTVGLDRDVNFYKLVSEKSDVNIVAGTGFYVEDVQAKANLSKTTEQMYHHMLTELTTGFLEEPSVKAGFMGEIASVWPLREFERRAIVAAGELQPQVGCGVSFHPHRAIEAPFEIIRLYLEAGGSKDKVVMSHLDRTLLEDDKLLEFSELGTYCQFDLFGVEVSFYQLNPAADMPSDGQRIDKIKLLIDDGKSDKVLMSHDIHTKHRLTAFGGHGYAHIINTVLPKMALKGISQDTIDRITIDNPARWLATNI
Summary
Cofactor
a divalent metal cation
Similarity
Belongs to the metallo-dependent hydrolases superfamily. Phosphotriesterase family.
Keywords
Complete proteome
Hydrolase
Metal-binding
Reference proteome
Feature
chain Phosphotriesterase-related protein
Uniprot
E5LAN2
A0A0N1PFY7
A0A2W1BUM2
F4YBG6
A0A2A4JLL2
A0A2H1W371
+ More
A0A194PW10 A0A212FK58 A0A2J7PTZ8 A0A1B6KBM8 A0A1B6K5Q2 A0A1B6ET17 A0A232EWT1 A0A1L8DCT3 A0A1L8DCJ6 A0A0P4VSL6 R4FNE8 A0A069DT59 A0A0L7LP92 T1PIE4 A0A0A1X022 A0A1L8EH97 A0A1L8EHA5 A0A1I8MAW5 A0A2A3E4M5 A0A088AAL0 A0A023F7R4 A0A0C9Q1N8 A0A2P8ZKG3 A0A0L0CNN8 A0A034VPI0 A0A1I8NPF1 A0A084WLX9 A0A182QZE6 A0A182V6D6 A0A1A9WQ53 A0A182HW22 A0A1L8EB29 A0A1L8EB80 U5EW16 A0A182KKP2 A0A182JK99 A0A154P1B0 A0A182U420 A0A224XRT4 A0A1B0C3F1 A0A182WUF6 W8BM77 A0A1B0FEJ0 B4J340 A0A1A9YST3 A0A1W4VQ91 B3M070 A0A026X1Z9 Q9VHF2 A0A182YBH7 A0A182N3T6 B4K4Y6 A0A182WDT7 Q7QK30 B4PUM2 A0A182PHA8 B4HKF2 B4NAJ1 A0A182RUJ4 A0A3B0JFQ2 A0A182JS72 A0A182M2R0 B3P1R1 A0A1J1HH38 B4LW09 A0A182FUP0 B4GP48 Q29BL3 A0A0M4EJL1 A0A067R7K6 E2BBZ2 T1DK56 E9IM59 W5JT27 A0A195EE76 E2AD64 A0A1A9ZYY3 A0A151IHF8 A0A195BWH7 F4WX19 A0A158NCH5 A0A151WMR2 A0A0A9WN84 G8Z4I4 A0A336LR71 A0A146M575 A0A1Q3G4X6 Q0IEH7 A0A0K8W1R8 A0A0N7ZCM4 A0A3R7LYV0 A0A1B6KYG9 A0A1A9VG18 A0A2P2I146
A0A194PW10 A0A212FK58 A0A2J7PTZ8 A0A1B6KBM8 A0A1B6K5Q2 A0A1B6ET17 A0A232EWT1 A0A1L8DCT3 A0A1L8DCJ6 A0A0P4VSL6 R4FNE8 A0A069DT59 A0A0L7LP92 T1PIE4 A0A0A1X022 A0A1L8EH97 A0A1L8EHA5 A0A1I8MAW5 A0A2A3E4M5 A0A088AAL0 A0A023F7R4 A0A0C9Q1N8 A0A2P8ZKG3 A0A0L0CNN8 A0A034VPI0 A0A1I8NPF1 A0A084WLX9 A0A182QZE6 A0A182V6D6 A0A1A9WQ53 A0A182HW22 A0A1L8EB29 A0A1L8EB80 U5EW16 A0A182KKP2 A0A182JK99 A0A154P1B0 A0A182U420 A0A224XRT4 A0A1B0C3F1 A0A182WUF6 W8BM77 A0A1B0FEJ0 B4J340 A0A1A9YST3 A0A1W4VQ91 B3M070 A0A026X1Z9 Q9VHF2 A0A182YBH7 A0A182N3T6 B4K4Y6 A0A182WDT7 Q7QK30 B4PUM2 A0A182PHA8 B4HKF2 B4NAJ1 A0A182RUJ4 A0A3B0JFQ2 A0A182JS72 A0A182M2R0 B3P1R1 A0A1J1HH38 B4LW09 A0A182FUP0 B4GP48 Q29BL3 A0A0M4EJL1 A0A067R7K6 E2BBZ2 T1DK56 E9IM59 W5JT27 A0A195EE76 E2AD64 A0A1A9ZYY3 A0A151IHF8 A0A195BWH7 F4WX19 A0A158NCH5 A0A151WMR2 A0A0A9WN84 G8Z4I4 A0A336LR71 A0A146M575 A0A1Q3G4X6 Q0IEH7 A0A0K8W1R8 A0A0N7ZCM4 A0A3R7LYV0 A0A1B6KYG9 A0A1A9VG18 A0A2P2I146
EC Number
3.1.-.-
Pubmed
19121390
26354079
28756777
22118469
28648823
27129103
+ More
26334808 26227816 25830018 25315136 25474469 29403074 26108605 25348373 24438588 20966253 24495485 17994087 24508170 30249741 10731132 12537572 12537569 25244985 12364791 15632085 24845553 20798317 21282665 20920257 23761445 21719571 21347285 25401762 26823975 17510324
26334808 26227816 25830018 25315136 25474469 29403074 26108605 25348373 24438588 20966253 24495485 17994087 24508170 30249741 10731132 12537572 12537569 25244985 12364791 15632085 24845553 20798317 21282665 20920257 23761445 21719571 21347285 25401762 26823975 17510324
EMBL
BABH01012392
HQ391899
ADP88941.1
KQ460971
KPJ10129.1
KZ149896
+ More
PZC78748.1 GU727738 ADK73626.1 NWSH01001090 PCG72666.1 ODYU01006045 SOQ47545.1 KQ459590 KPI97183.1 AGBW02008141 OWR54121.1 NEVH01021216 PNF19815.1 GEBQ01031141 JAT08836.1 GECU01000931 JAT06776.1 GECZ01028670 JAS41099.1 NNAY01001823 OXU22807.1 GFDF01009907 JAV04177.1 GFDF01009908 JAV04176.1 GDKW01001644 JAI54951.1 ACPB03001919 GAHY01001324 JAA76186.1 GBGD01001988 JAC86901.1 JTDY01000437 KOB77174.1 KA647698 AFP62327.1 GBXI01010061 JAD04231.1 GFDG01000765 JAV18034.1 GFDG01000764 JAV18035.1 KZ288377 PBC26650.1 GBBI01001439 JAC17273.1 GBYB01007833 JAG77600.1 PYGN01000029 PSN56987.1 JRES01000145 KNC33842.1 GAKP01014568 JAC44384.1 ATLV01024321 KE525351 KFB51223.1 AXCN02001098 APCN01001457 GFDG01002935 JAV15864.1 GFDG01002934 JAV15865.1 GANO01001638 JAB58233.1 KQ434786 KZC05134.1 GFTR01005705 JAW10721.1 JXJN01024937 GAMC01004350 JAC02206.1 CCAG010001988 CH916366 CH902617 KK107034 QOIP01000008 EZA62026.1 RLU19607.1 AE014297 AY071616 CH933806 AAAB01008804 EAA03848.2 CM000160 CH480815 CH964232 OUUW01000005 SPP81167.1 AXCM01001194 CH954181 CVRI01000004 CRK87335.1 CH940650 CH479186 CM000070 CP012526 ALC47134.1 KK852649 KDR19492.1 GL447175 EFN86816.1 GAMD01001111 JAB00480.1 GL764129 EFZ18317.1 ADMH02000555 ETN65894.1 KQ979074 KYN23102.1 GL438684 EFN68612.1 KQ977613 KYN01394.1 KQ976396 KYM92937.1 GL888417 EGI61262.1 ADTU01011886 KQ982934 KYQ49192.1 GBHO01037289 JAG06315.1 GU123165 ADK62518.1 UFQS01000081 UFQT01000081 SSW99049.1 SSX19431.1 GDHC01004467 JAQ14162.1 GFDL01000229 JAV34816.1 CH477655 GDHF01007293 JAI45021.1 GDRN01064334 JAI64856.1 QCYY01002871 ROT66984.1 GEBQ01023495 JAT16482.1 IACF01002087 LAB67755.1
PZC78748.1 GU727738 ADK73626.1 NWSH01001090 PCG72666.1 ODYU01006045 SOQ47545.1 KQ459590 KPI97183.1 AGBW02008141 OWR54121.1 NEVH01021216 PNF19815.1 GEBQ01031141 JAT08836.1 GECU01000931 JAT06776.1 GECZ01028670 JAS41099.1 NNAY01001823 OXU22807.1 GFDF01009907 JAV04177.1 GFDF01009908 JAV04176.1 GDKW01001644 JAI54951.1 ACPB03001919 GAHY01001324 JAA76186.1 GBGD01001988 JAC86901.1 JTDY01000437 KOB77174.1 KA647698 AFP62327.1 GBXI01010061 JAD04231.1 GFDG01000765 JAV18034.1 GFDG01000764 JAV18035.1 KZ288377 PBC26650.1 GBBI01001439 JAC17273.1 GBYB01007833 JAG77600.1 PYGN01000029 PSN56987.1 JRES01000145 KNC33842.1 GAKP01014568 JAC44384.1 ATLV01024321 KE525351 KFB51223.1 AXCN02001098 APCN01001457 GFDG01002935 JAV15864.1 GFDG01002934 JAV15865.1 GANO01001638 JAB58233.1 KQ434786 KZC05134.1 GFTR01005705 JAW10721.1 JXJN01024937 GAMC01004350 JAC02206.1 CCAG010001988 CH916366 CH902617 KK107034 QOIP01000008 EZA62026.1 RLU19607.1 AE014297 AY071616 CH933806 AAAB01008804 EAA03848.2 CM000160 CH480815 CH964232 OUUW01000005 SPP81167.1 AXCM01001194 CH954181 CVRI01000004 CRK87335.1 CH940650 CH479186 CM000070 CP012526 ALC47134.1 KK852649 KDR19492.1 GL447175 EFN86816.1 GAMD01001111 JAB00480.1 GL764129 EFZ18317.1 ADMH02000555 ETN65894.1 KQ979074 KYN23102.1 GL438684 EFN68612.1 KQ977613 KYN01394.1 KQ976396 KYM92937.1 GL888417 EGI61262.1 ADTU01011886 KQ982934 KYQ49192.1 GBHO01037289 JAG06315.1 GU123165 ADK62518.1 UFQS01000081 UFQT01000081 SSW99049.1 SSX19431.1 GDHC01004467 JAQ14162.1 GFDL01000229 JAV34816.1 CH477655 GDHF01007293 JAI45021.1 GDRN01064334 JAI64856.1 QCYY01002871 ROT66984.1 GEBQ01023495 JAT16482.1 IACF01002087 LAB67755.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000235965
+ More
UP000215335 UP000015103 UP000037510 UP000095301 UP000242457 UP000005203 UP000245037 UP000037069 UP000095300 UP000030765 UP000075886 UP000075903 UP000091820 UP000075840 UP000075882 UP000075880 UP000076502 UP000075902 UP000092460 UP000076407 UP000092444 UP000001070 UP000092443 UP000192221 UP000007801 UP000053097 UP000279307 UP000000803 UP000076408 UP000075884 UP000009192 UP000075920 UP000007062 UP000002282 UP000075885 UP000001292 UP000007798 UP000075900 UP000268350 UP000075881 UP000075883 UP000008711 UP000183832 UP000008792 UP000069272 UP000008744 UP000001819 UP000092553 UP000027135 UP000008237 UP000000673 UP000078492 UP000000311 UP000092445 UP000078542 UP000078540 UP000007755 UP000005205 UP000075809 UP000008820 UP000283509 UP000078200
UP000215335 UP000015103 UP000037510 UP000095301 UP000242457 UP000005203 UP000245037 UP000037069 UP000095300 UP000030765 UP000075886 UP000075903 UP000091820 UP000075840 UP000075882 UP000075880 UP000076502 UP000075902 UP000092460 UP000076407 UP000092444 UP000001070 UP000092443 UP000192221 UP000007801 UP000053097 UP000279307 UP000000803 UP000076408 UP000075884 UP000009192 UP000075920 UP000007062 UP000002282 UP000075885 UP000001292 UP000007798 UP000075900 UP000268350 UP000075881 UP000075883 UP000008711 UP000183832 UP000008792 UP000069272 UP000008744 UP000001819 UP000092553 UP000027135 UP000008237 UP000000673 UP000078492 UP000000311 UP000092445 UP000078542 UP000078540 UP000007755 UP000005205 UP000075809 UP000008820 UP000283509 UP000078200
Pfam
PF02126 PTE
Interpro
SUPFAM
SSF51556
SSF51556
CDD
ProteinModelPortal
E5LAN2
A0A0N1PFY7
A0A2W1BUM2
F4YBG6
A0A2A4JLL2
A0A2H1W371
+ More
A0A194PW10 A0A212FK58 A0A2J7PTZ8 A0A1B6KBM8 A0A1B6K5Q2 A0A1B6ET17 A0A232EWT1 A0A1L8DCT3 A0A1L8DCJ6 A0A0P4VSL6 R4FNE8 A0A069DT59 A0A0L7LP92 T1PIE4 A0A0A1X022 A0A1L8EH97 A0A1L8EHA5 A0A1I8MAW5 A0A2A3E4M5 A0A088AAL0 A0A023F7R4 A0A0C9Q1N8 A0A2P8ZKG3 A0A0L0CNN8 A0A034VPI0 A0A1I8NPF1 A0A084WLX9 A0A182QZE6 A0A182V6D6 A0A1A9WQ53 A0A182HW22 A0A1L8EB29 A0A1L8EB80 U5EW16 A0A182KKP2 A0A182JK99 A0A154P1B0 A0A182U420 A0A224XRT4 A0A1B0C3F1 A0A182WUF6 W8BM77 A0A1B0FEJ0 B4J340 A0A1A9YST3 A0A1W4VQ91 B3M070 A0A026X1Z9 Q9VHF2 A0A182YBH7 A0A182N3T6 B4K4Y6 A0A182WDT7 Q7QK30 B4PUM2 A0A182PHA8 B4HKF2 B4NAJ1 A0A182RUJ4 A0A3B0JFQ2 A0A182JS72 A0A182M2R0 B3P1R1 A0A1J1HH38 B4LW09 A0A182FUP0 B4GP48 Q29BL3 A0A0M4EJL1 A0A067R7K6 E2BBZ2 T1DK56 E9IM59 W5JT27 A0A195EE76 E2AD64 A0A1A9ZYY3 A0A151IHF8 A0A195BWH7 F4WX19 A0A158NCH5 A0A151WMR2 A0A0A9WN84 G8Z4I4 A0A336LR71 A0A146M575 A0A1Q3G4X6 Q0IEH7 A0A0K8W1R8 A0A0N7ZCM4 A0A3R7LYV0 A0A1B6KYG9 A0A1A9VG18 A0A2P2I146
A0A194PW10 A0A212FK58 A0A2J7PTZ8 A0A1B6KBM8 A0A1B6K5Q2 A0A1B6ET17 A0A232EWT1 A0A1L8DCT3 A0A1L8DCJ6 A0A0P4VSL6 R4FNE8 A0A069DT59 A0A0L7LP92 T1PIE4 A0A0A1X022 A0A1L8EH97 A0A1L8EHA5 A0A1I8MAW5 A0A2A3E4M5 A0A088AAL0 A0A023F7R4 A0A0C9Q1N8 A0A2P8ZKG3 A0A0L0CNN8 A0A034VPI0 A0A1I8NPF1 A0A084WLX9 A0A182QZE6 A0A182V6D6 A0A1A9WQ53 A0A182HW22 A0A1L8EB29 A0A1L8EB80 U5EW16 A0A182KKP2 A0A182JK99 A0A154P1B0 A0A182U420 A0A224XRT4 A0A1B0C3F1 A0A182WUF6 W8BM77 A0A1B0FEJ0 B4J340 A0A1A9YST3 A0A1W4VQ91 B3M070 A0A026X1Z9 Q9VHF2 A0A182YBH7 A0A182N3T6 B4K4Y6 A0A182WDT7 Q7QK30 B4PUM2 A0A182PHA8 B4HKF2 B4NAJ1 A0A182RUJ4 A0A3B0JFQ2 A0A182JS72 A0A182M2R0 B3P1R1 A0A1J1HH38 B4LW09 A0A182FUP0 B4GP48 Q29BL3 A0A0M4EJL1 A0A067R7K6 E2BBZ2 T1DK56 E9IM59 W5JT27 A0A195EE76 E2AD64 A0A1A9ZYY3 A0A151IHF8 A0A195BWH7 F4WX19 A0A158NCH5 A0A151WMR2 A0A0A9WN84 G8Z4I4 A0A336LR71 A0A146M575 A0A1Q3G4X6 Q0IEH7 A0A0K8W1R8 A0A0N7ZCM4 A0A3R7LYV0 A0A1B6KYG9 A0A1A9VG18 A0A2P2I146
PDB
3K2G
E-value=3.43364e-32,
Score=345
Ontologies
GO
PANTHER
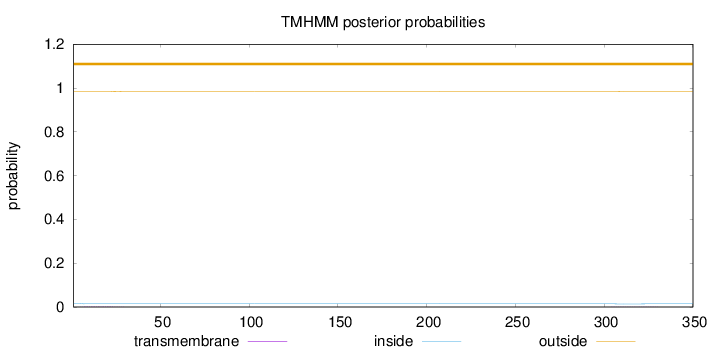
Topology
Length:
350
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0541900000000001
Exp number, first 60 AAs:
0.0362
Total prob of N-in:
0.01618
outside
1 - 350
Population Genetic Test Statistics
Pi
18.384139
Theta
177.47815
Tajima's D
0.375363
CLR
1.385852
CSRT
0.47667616619169
Interpretation
Uncertain
