Gene
KWMTBOMO06433
Pre Gene Modal
BGIBMGA011881
Annotation
PREDICTED:_transmembrane_protein_64-like_isoform_X1_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 4.987
Sequence
CDS
ATGGAAATCGTTGACATCGAAGAATGCCAGCCCGACCCTGTTGCGGTCACTAGAAAGAAAAGTTTATGGTCTAAAATTAACAACAAAACAACTTACAGTTACTTGTTCAATATAGTTATATCAATACTTTTATTAACTTGTCTCGTTTTGGTGTTGTACTTTTTTAAAGAATATCTAAAGACTGTTTTATATTGGGTCGACGCTCAAGATCCGTGGATTATTTTTTTGTTGTTCATGTGTCTGTTTCTGATCGTGAGTTTTCCGGTGGCTGTTGGTTATTTAATTTTGATAATAACGAGCGGCTATCTGTTCGGGATGCTGAAGGGCATGGGAACTGTGTTAGTGAGTGCTAATTTTGGCGTGGCGGTCGCTCATTTCACGTTGAAGGCGTTAAGAGCGTATCTTCCACTTGACAAACTGTTAAAGCATGACACCGCAAGAGCTCTGCTCAAGGTTATCTCGGGACCTCAAGCTTTTAAGATTGTGTTCTTCGCGAGACTCACGCCGATACCTTTTGGATTGCAGAACACTATTTTCGCTGTAAGCGACGTTCACGGTTGCGGCTACCATCTCGCGACGATGCTCGGTCTGCTGCCGGCGCAGGTGATCAATGTCTACCTCGGCTCGACGCTCCGGTCCATCCACGACGTGCTCCACGAGTCCCACGTCACCGGATACATCGTGTTCGCTTTCCAGATATTAATCGGCATTACACTGATGGTGTGGGTCGTGCAAAAAGCCCGAAAAGAGCTCACCATAGCGATAATGGCCGCGGAACTAGGCCGAGAAACAATATCGACGTCAAGCAGCTCCTCGATCAGCTAG
Protein
MEIVDIEECQPDPVAVTRKKSLWSKINNKTTYSYLFNIVISILLLTCLVLVLYFFKEYLKTVLYWVDAQDPWIIFLLFMCLFLIVSFPVAVGYLILIITSGYLFGMLKGMGTVLVSANFGVAVAHFTLKALRAYLPLDKLLKHDTARALLKVISGPQAFKIVFFARLTPIPFGLQNTIFAVSDVHGCGYHLATMLGLLPAQVINVYLGSTLRSIHDVLHESHVTGYIVFAFQILIGITLMVWVVQKARKELTIAIMAAELGRETISTSSSSSIS
Summary
Uniprot
A0A2W1C1A1
A0A2H1WHI1
A0A1E1W1Y1
A0A2A4JM56
A0A154P2K0
A0A0N0BGK0
+ More
A0A0L7QU66 A0A0K8TKI2 A0A088A1W9 A0A2A3ENE3 K7IYE2 V9IKV5 A0A026WFR1 A0A151X1V6 E9J4J8 F4WK21 A0A151IEA1 A0A195BWS4 A0A158NM68 E2ABV7 T1DEC8 A0A182Y1G7 A0A1Q3EUB0 A0A232EL61 A0A2M4AQ18 A0A0L0C8B1 A0A0P9AMS8 A0A3B0J484 W5JKD7 A0A2M3ZGL3 A0A2M3Z382 A0A2M3Z3N2 A0A2M4AKR6 A0A1W4URA4 A0A2M4BNG4 Q5TMY2 A0A2M4BNQ3 A0A2M4BNI1 A0A2M4ALN6 A0A2M4BNN1 B3MB81 B4N5G1 A0A182P5V4 A0A182FBC3 A0A182WEX5 A0A182Q9E3 Q17NM9 A0A0J9S018 A0A084WIX1 B4PIP7 Q29F14 Q9VNR8 B4HC16 T1PIT8 A0A1I8M7K0 A0A1I8PFU0 A0A1A9WD86 A0A023ESA1 A0A182WZT8 B3NEF2 A0A2C9GR00 A0A182MGN8 W8BEU2 A0A182UVU9 A0A1B0B6X8 A0A1A9VFA5 A0A034VL26 A0A182JTP7 B4J3Q2 B4KYJ2 B4LDY3 A0A182ISM4 A0A182RD12 A0A0A1WSB1 A0A034VPH2 A0A0K8VRB6 E2B3S3 A0A1J1IC11 A0A336K7M3 A0A151JVI4 E0VVW3 A0A1A9Y0Y6 A0A182KSB3 A0A1W4XB57 A0A067RKU2 E9GEQ8 A0A164YDZ5 A0A0T6B757 A0A1Y1KMU5 A0A3B3UEA5 A0A3B5MYB6 A0A3B5QQF3
A0A0L7QU66 A0A0K8TKI2 A0A088A1W9 A0A2A3ENE3 K7IYE2 V9IKV5 A0A026WFR1 A0A151X1V6 E9J4J8 F4WK21 A0A151IEA1 A0A195BWS4 A0A158NM68 E2ABV7 T1DEC8 A0A182Y1G7 A0A1Q3EUB0 A0A232EL61 A0A2M4AQ18 A0A0L0C8B1 A0A0P9AMS8 A0A3B0J484 W5JKD7 A0A2M3ZGL3 A0A2M3Z382 A0A2M3Z3N2 A0A2M4AKR6 A0A1W4URA4 A0A2M4BNG4 Q5TMY2 A0A2M4BNQ3 A0A2M4BNI1 A0A2M4ALN6 A0A2M4BNN1 B3MB81 B4N5G1 A0A182P5V4 A0A182FBC3 A0A182WEX5 A0A182Q9E3 Q17NM9 A0A0J9S018 A0A084WIX1 B4PIP7 Q29F14 Q9VNR8 B4HC16 T1PIT8 A0A1I8M7K0 A0A1I8PFU0 A0A1A9WD86 A0A023ESA1 A0A182WZT8 B3NEF2 A0A2C9GR00 A0A182MGN8 W8BEU2 A0A182UVU9 A0A1B0B6X8 A0A1A9VFA5 A0A034VL26 A0A182JTP7 B4J3Q2 B4KYJ2 B4LDY3 A0A182ISM4 A0A182RD12 A0A0A1WSB1 A0A034VPH2 A0A0K8VRB6 E2B3S3 A0A1J1IC11 A0A336K7M3 A0A151JVI4 E0VVW3 A0A1A9Y0Y6 A0A182KSB3 A0A1W4XB57 A0A067RKU2 E9GEQ8 A0A164YDZ5 A0A0T6B757 A0A1Y1KMU5 A0A3B3UEA5 A0A3B5MYB6 A0A3B5QQF3
Pubmed
28756777
26369729
20075255
24508170
30249741
21282665
+ More
21719571 21347285 20798317 24330624 25244985 28648823 26108605 17994087 20920257 23761445 12364791 17510324 22936249 24438588 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 24945155 24495485 25348373 25830018 20566863 20966253 24845553 21292972 28004739 23542700
21719571 21347285 20798317 24330624 25244985 28648823 26108605 17994087 20920257 23761445 12364791 17510324 22936249 24438588 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 24945155 24495485 25348373 25830018 20566863 20966253 24845553 21292972 28004739 23542700
EMBL
KZ149896
PZC78746.1
ODYU01008710
SOQ52523.1
GDQN01010057
JAT80997.1
+ More
NWSH01001090 PCG72664.1 KQ434792 KZC05448.1 KQ435776 KOX74912.1 KQ414735 KOC62202.1 GDAI01002992 JAI14611.1 KZ288215 PBC32659.1 JR050590 AEY61312.1 KK107238 QOIP01000007 EZA54773.1 RLU21084.1 KQ982584 KYQ54396.1 GL768105 EFZ12230.1 GL888190 EGI65471.1 KQ977890 KYM98990.1 KQ976395 KYM93084.1 ADTU01020317 ADTU01020318 GL438386 EFN69052.1 GALA01001087 JAA93765.1 GFDL01016145 JAV18900.1 NNAY01003621 OXU19094.1 GGFK01009553 MBW42874.1 JRES01000763 KNC28485.1 CH902618 KPU79079.1 OUUW01000002 SPP76297.1 ADMH02001279 ETN63239.1 GGFM01006935 MBW27686.1 GGFM01002223 MBW22974.1 GGFM01002380 MBW23131.1 GGFK01008065 MBW41386.1 GGFJ01005485 MBW54626.1 AAAB01008986 EAL38667.3 GGFJ01005430 MBW54571.1 GGFJ01005431 MBW54572.1 GGFK01008321 MBW41642.1 GGFJ01005432 MBW54573.1 EDV41382.1 CH964101 EDW79600.1 AXCN02001025 CH477198 EAT48284.1 CM002912 KMZ01219.1 ATLV01023952 KE525347 KFB50165.1 CM000159 EDW95555.1 CH379070 EAL29895.2 AE014296 AY052011 AAF51854.3 AAK93435.1 CH479275 EDW40047.1 KA648722 AFP63351.1 GAPW01001982 JAC11616.1 CH954178 EDV52787.1 APCN01003309 AXCM01006675 GAMC01014809 JAB91746.1 JXJN01009256 JXJN01009257 JXJN01009258 GAKP01014931 JAC44021.1 CH916366 EDV97283.1 CH933809 EDW18803.2 CH940647 EDW70026.2 GBXI01012907 GBXI01008838 JAD01385.1 JAD05454.1 GAKP01014930 GAKP01014929 JAC44022.1 GDHF01010936 JAI41378.1 GL445398 EFN89660.1 CVRI01000047 CRK97739.1 UFQS01000187 UFQT01000187 SSX00956.1 SSX21336.1 KQ981697 KYN37534.1 DS235815 EEB17519.1 KK852567 KDR21205.1 GL732541 EFX82037.1 LRGB01000915 KZS15161.1 LJIG01009392 KRT83171.1 GEZM01078431 JAV62722.1
NWSH01001090 PCG72664.1 KQ434792 KZC05448.1 KQ435776 KOX74912.1 KQ414735 KOC62202.1 GDAI01002992 JAI14611.1 KZ288215 PBC32659.1 JR050590 AEY61312.1 KK107238 QOIP01000007 EZA54773.1 RLU21084.1 KQ982584 KYQ54396.1 GL768105 EFZ12230.1 GL888190 EGI65471.1 KQ977890 KYM98990.1 KQ976395 KYM93084.1 ADTU01020317 ADTU01020318 GL438386 EFN69052.1 GALA01001087 JAA93765.1 GFDL01016145 JAV18900.1 NNAY01003621 OXU19094.1 GGFK01009553 MBW42874.1 JRES01000763 KNC28485.1 CH902618 KPU79079.1 OUUW01000002 SPP76297.1 ADMH02001279 ETN63239.1 GGFM01006935 MBW27686.1 GGFM01002223 MBW22974.1 GGFM01002380 MBW23131.1 GGFK01008065 MBW41386.1 GGFJ01005485 MBW54626.1 AAAB01008986 EAL38667.3 GGFJ01005430 MBW54571.1 GGFJ01005431 MBW54572.1 GGFK01008321 MBW41642.1 GGFJ01005432 MBW54573.1 EDV41382.1 CH964101 EDW79600.1 AXCN02001025 CH477198 EAT48284.1 CM002912 KMZ01219.1 ATLV01023952 KE525347 KFB50165.1 CM000159 EDW95555.1 CH379070 EAL29895.2 AE014296 AY052011 AAF51854.3 AAK93435.1 CH479275 EDW40047.1 KA648722 AFP63351.1 GAPW01001982 JAC11616.1 CH954178 EDV52787.1 APCN01003309 AXCM01006675 GAMC01014809 JAB91746.1 JXJN01009256 JXJN01009257 JXJN01009258 GAKP01014931 JAC44021.1 CH916366 EDV97283.1 CH933809 EDW18803.2 CH940647 EDW70026.2 GBXI01012907 GBXI01008838 JAD01385.1 JAD05454.1 GAKP01014930 GAKP01014929 JAC44022.1 GDHF01010936 JAI41378.1 GL445398 EFN89660.1 CVRI01000047 CRK97739.1 UFQS01000187 UFQT01000187 SSX00956.1 SSX21336.1 KQ981697 KYN37534.1 DS235815 EEB17519.1 KK852567 KDR21205.1 GL732541 EFX82037.1 LRGB01000915 KZS15161.1 LJIG01009392 KRT83171.1 GEZM01078431 JAV62722.1
Proteomes
UP000218220
UP000076502
UP000053105
UP000053825
UP000005203
UP000242457
+ More
UP000002358 UP000053097 UP000279307 UP000075809 UP000007755 UP000078542 UP000078540 UP000005205 UP000000311 UP000076408 UP000215335 UP000037069 UP000007801 UP000268350 UP000000673 UP000192221 UP000007062 UP000007798 UP000075885 UP000069272 UP000075920 UP000075886 UP000008820 UP000030765 UP000002282 UP000001819 UP000000803 UP000008744 UP000095301 UP000095300 UP000091820 UP000076407 UP000008711 UP000075840 UP000075883 UP000075903 UP000092460 UP000078200 UP000075881 UP000001070 UP000009192 UP000008792 UP000075880 UP000075900 UP000008237 UP000183832 UP000078541 UP000009046 UP000092443 UP000075882 UP000192223 UP000027135 UP000000305 UP000076858 UP000261500 UP000261380 UP000002852
UP000002358 UP000053097 UP000279307 UP000075809 UP000007755 UP000078542 UP000078540 UP000005205 UP000000311 UP000076408 UP000215335 UP000037069 UP000007801 UP000268350 UP000000673 UP000192221 UP000007062 UP000007798 UP000075885 UP000069272 UP000075920 UP000075886 UP000008820 UP000030765 UP000002282 UP000001819 UP000000803 UP000008744 UP000095301 UP000095300 UP000091820 UP000076407 UP000008711 UP000075840 UP000075883 UP000075903 UP000092460 UP000078200 UP000075881 UP000001070 UP000009192 UP000008792 UP000075880 UP000075900 UP000008237 UP000183832 UP000078541 UP000009046 UP000092443 UP000075882 UP000192223 UP000027135 UP000000305 UP000076858 UP000261500 UP000261380 UP000002852
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A0A2W1C1A1
A0A2H1WHI1
A0A1E1W1Y1
A0A2A4JM56
A0A154P2K0
A0A0N0BGK0
+ More
A0A0L7QU66 A0A0K8TKI2 A0A088A1W9 A0A2A3ENE3 K7IYE2 V9IKV5 A0A026WFR1 A0A151X1V6 E9J4J8 F4WK21 A0A151IEA1 A0A195BWS4 A0A158NM68 E2ABV7 T1DEC8 A0A182Y1G7 A0A1Q3EUB0 A0A232EL61 A0A2M4AQ18 A0A0L0C8B1 A0A0P9AMS8 A0A3B0J484 W5JKD7 A0A2M3ZGL3 A0A2M3Z382 A0A2M3Z3N2 A0A2M4AKR6 A0A1W4URA4 A0A2M4BNG4 Q5TMY2 A0A2M4BNQ3 A0A2M4BNI1 A0A2M4ALN6 A0A2M4BNN1 B3MB81 B4N5G1 A0A182P5V4 A0A182FBC3 A0A182WEX5 A0A182Q9E3 Q17NM9 A0A0J9S018 A0A084WIX1 B4PIP7 Q29F14 Q9VNR8 B4HC16 T1PIT8 A0A1I8M7K0 A0A1I8PFU0 A0A1A9WD86 A0A023ESA1 A0A182WZT8 B3NEF2 A0A2C9GR00 A0A182MGN8 W8BEU2 A0A182UVU9 A0A1B0B6X8 A0A1A9VFA5 A0A034VL26 A0A182JTP7 B4J3Q2 B4KYJ2 B4LDY3 A0A182ISM4 A0A182RD12 A0A0A1WSB1 A0A034VPH2 A0A0K8VRB6 E2B3S3 A0A1J1IC11 A0A336K7M3 A0A151JVI4 E0VVW3 A0A1A9Y0Y6 A0A182KSB3 A0A1W4XB57 A0A067RKU2 E9GEQ8 A0A164YDZ5 A0A0T6B757 A0A1Y1KMU5 A0A3B3UEA5 A0A3B5MYB6 A0A3B5QQF3
A0A0L7QU66 A0A0K8TKI2 A0A088A1W9 A0A2A3ENE3 K7IYE2 V9IKV5 A0A026WFR1 A0A151X1V6 E9J4J8 F4WK21 A0A151IEA1 A0A195BWS4 A0A158NM68 E2ABV7 T1DEC8 A0A182Y1G7 A0A1Q3EUB0 A0A232EL61 A0A2M4AQ18 A0A0L0C8B1 A0A0P9AMS8 A0A3B0J484 W5JKD7 A0A2M3ZGL3 A0A2M3Z382 A0A2M3Z3N2 A0A2M4AKR6 A0A1W4URA4 A0A2M4BNG4 Q5TMY2 A0A2M4BNQ3 A0A2M4BNI1 A0A2M4ALN6 A0A2M4BNN1 B3MB81 B4N5G1 A0A182P5V4 A0A182FBC3 A0A182WEX5 A0A182Q9E3 Q17NM9 A0A0J9S018 A0A084WIX1 B4PIP7 Q29F14 Q9VNR8 B4HC16 T1PIT8 A0A1I8M7K0 A0A1I8PFU0 A0A1A9WD86 A0A023ESA1 A0A182WZT8 B3NEF2 A0A2C9GR00 A0A182MGN8 W8BEU2 A0A182UVU9 A0A1B0B6X8 A0A1A9VFA5 A0A034VL26 A0A182JTP7 B4J3Q2 B4KYJ2 B4LDY3 A0A182ISM4 A0A182RD12 A0A0A1WSB1 A0A034VPH2 A0A0K8VRB6 E2B3S3 A0A1J1IC11 A0A336K7M3 A0A151JVI4 E0VVW3 A0A1A9Y0Y6 A0A182KSB3 A0A1W4XB57 A0A067RKU2 E9GEQ8 A0A164YDZ5 A0A0T6B757 A0A1Y1KMU5 A0A3B3UEA5 A0A3B5MYB6 A0A3B5QQF3
Ontologies
PANTHER
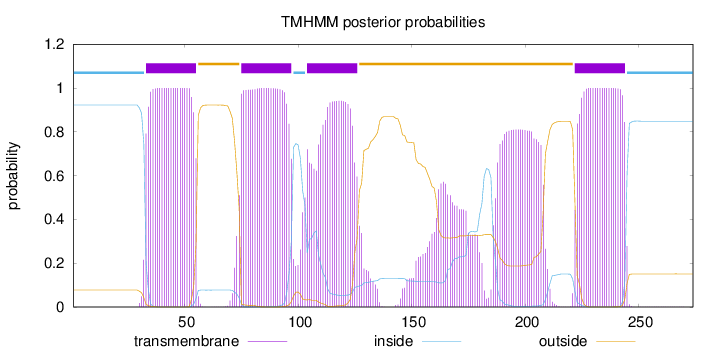
Topology
Length:
274
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
120.54152
Exp number, first 60 AAs:
22.55374
Total prob of N-in:
0.92261
POSSIBLE N-term signal
sequence
inside
1 - 32
TMhelix
33 - 55
outside
56 - 74
TMhelix
75 - 97
inside
98 - 103
TMhelix
104 - 126
outside
127 - 221
TMhelix
222 - 244
inside
245 - 274
Population Genetic Test Statistics
Pi
251.724568
Theta
167.354219
Tajima's D
1.253016
CLR
0.342048
CSRT
0.724863756812159
Interpretation
Uncertain
