Gene
KWMTBOMO06428 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011601
Annotation
Alanine--glyoxylate_aminotransferase_2?_mitochondrial_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.451 Mitochondrial Reliability : 1.012
Sequence
CDS
ATGGCCAATAGAGGACGACTGTGCAATGTGGTTGGCAGGGCGTATAGTACAGCTAAAATGCCCTCCACCGGGTTTACACCCAAAGAGTATAAGGGCCCAACATACGAACAACTGGAACATTTGAAGTCGAAACATATGCCGCCAGCGATACCTAATTCTTACAAGAAGCCGGTGCTATTACACCAGGGTCACATGCAGTGGCTCTTCGACCACGAGGGCAAAAGATACTTGGACCTGTTCGGCGGAGTCGTTACTGTCTCAGTTGGAAATAGTCATCCGAAGCTAAACGCAGCTCTCAAAGAACAAATCGATTTAATATGGCACACGACGAACTTGTACCGGCATCCAAGAATATATGAATACATTGAGAAATTGTCGTCTAAATTACCCGATGATCTTAACGTCGTCTACTTAGTGAACAGTGGAACAGAAGCGAACGATCTGGCCACGATCCTGGCAAAGGCGTACACGGGTAACTTGGACGTGATATCACTCCAGACCAGCTATCATGGATACAACAGCAGCCTCATGGGGCTATCTGCTACACAGTCGTACAGAATGAACCTCCCCGTACCGCCGGGATTTTATCACGCGGCACATCCAGATCCGTACCGAGGGAGCTGGGGCGGCTGCAGAGACTCCATTTCGCAGGTGGCCGGCGCGTGCTCGTGTCCCGGCGAATGCGTTAGCACCGAAAAATACATTCATCAACTCGACGAGCTCCTAGGTAATTCTGTACCAGCCGGCAGAGTAGCAGCGTTCTTCGCGGAATCTATCCAGGGTGCTGGTGGAGTGGTGCAGTTCCCTAAGGGATACCTGAAGAGAGCTCAAGAGCTAATCAAGAAGAATGGCGGCTTGTATGTAGCTGATGAGGTCCAGACGGGTTTCGGTCGTACCGGAGACCACTACTGGGGCTTCGAGACACACGGCGTGAAGCCTGATATCGTGACGATGGCTAAGGGGATCGGCAACGGTTTCCCTCTGGCCGCTGTCGTCACCACTAAAGAGATAGCAGCCGCCCACAGCGGAAAATCGTATTTCAACACTTTTGGAGGAAACGCTTTGGCCGCAACAGTTGGAAAGGCCGTATTAGATGTTATTGACGAGGAAGAACTTCAAAAGAACAGCAAAACTGTTGGCGAATACTTCATCAGGCAGTTGATGGAGCTACAGAAGCAGCATCCAGTGATTGGCGATGTGAGAGGACAGGGCCTCATGATCGGGGTCGAGCTGGTCGAGCCTGGCACTAAGAAACCCCTGCCGGTGTCCCTTGTGAAAGATATACACGAAGAGATTAAAGACAATGGAGTTCTAGTCGTACTTGGCGGGCGGTGGAGTAATGTGTTCAGAATGAAGCCGCCGATGTGCATCACGAAGGCAGATGTTGATTTTGAGAAACACATCGAAATGGCTAATAGAGGAACAAAATTATGCTTCGACATCGTCAGAACGTACAGCACGGCCAAAATGCCGCCGACGGATTTCGTGCCCAGACCGTACACGGGACCTTCGTACCAACAAGTAGAACAAATGAAAGGAGTTTACATGCCGCCTTCAATCACGAACGCCTATAAGAAGCCGGTGCTTCTCACCCAGGGGCACATGCAGTGGTTATACGACAACGACGGCAAGAGATACCTGGATCTGTTCGGTGGAATCGTCACCGTCTCCGTGGGCCATTGTCATCCAAAAGTAAATGCAGCCCTCAAAGATCAACTCGATGTACTGTGGCATACGACCAACCTGTACAGACATCCGAAAATCTATGAGTACGTCGAACAATTAGCCGCTAAGCTCCCGGGGGATTTGAACGTGGTATATTTAGTGAACAGTGGATCAGAAGCTAATGAACTGGCGACTTTGCTGGCGAAAGCGTACACAGGAAACCTAGACATCATATCGCTGCAGACCAGCTACCACGGCTACACCAGCAGCCTCATGGGCCTGACGGCAACCCAGTCGTACAGGATGGCCATTCCCGTCCCTCCAGGTTTCTACCACGCAGTCCACCCTGATCCTTTCAGAGGCGCTTTCGGAGGCTGCAGGGACTCTATCTCGCAAGCTCCAGGATCCTGCTCGTGCACCGGAGAGTGCATCAGCACTGACAAATATGTCCACCAACTTAACGAGCTATTAGGTAACTCAGTTCCGGCCGGCAGAGTAGCGGCATTCTTTGCGGAATCCATTCAGGGTGCTGGTGGAGTCGTGCAATTCCCTAAGGGATACCTGAAGAAAGCTCAAGAGTTGATCAAGAAGAATGGTGGCTTGTATGTAGCTGATGAGGTACAAACAGGCTTCGGTCGCACTGGAGATCATTTCTGGGGCTTCGAGACGCACGGCGTGAAGCCCGACATCGTGACGATGGCTAAGGGGATCGGCAACGGATTCCCTCTGGCCGCTGTCGTCACCACTAAAGAGATAGCAGCCAATCACGCGAAGGCTGCCTACTTTAACACTTTCGGTGGAAACCCGATGGCTTCGACTGTTGGAAAAGCTGTATTGGAGGTTATTGAAGAAGAAGGACTGCAACAGAACAGTAAGGTAGTCGGCGAATACTTCATCAGGCAGTTGATGGATCTTCAGAAGCAGCACCCAGTGATTGGCGATGTCAGGGGACAAGGCCTCATGATCGGAGTGGAGCTTGTTGAACCCGGAACTAAGACACCGCTGACCACTTCCAAAGTTAATGATATCCATGAGAATATCAAAGACAACGGAGTGCTCATCGCTCGTGGTGGACGCTTTAATAATGTATTCAGAATAAAACCTCCAATGTGCATCACTAAACAGGACGTCGACTTCGGCATTTCGATCATCAATGATGCGATCAAAAAAGTTGTCAACTAA
Protein
MANRGRLCNVVGRAYSTAKMPSTGFTPKEYKGPTYEQLEHLKSKHMPPAIPNSYKKPVLLHQGHMQWLFDHEGKRYLDLFGGVVTVSVGNSHPKLNAALKEQIDLIWHTTNLYRHPRIYEYIEKLSSKLPDDLNVVYLVNSGTEANDLATILAKAYTGNLDVISLQTSYHGYNSSLMGLSATQSYRMNLPVPPGFYHAAHPDPYRGSWGGCRDSISQVAGACSCPGECVSTEKYIHQLDELLGNSVPAGRVAAFFAESIQGAGGVVQFPKGYLKRAQELIKKNGGLYVADEVQTGFGRTGDHYWGFETHGVKPDIVTMAKGIGNGFPLAAVVTTKEIAAAHSGKSYFNTFGGNALAATVGKAVLDVIDEEELQKNSKTVGEYFIRQLMELQKQHPVIGDVRGQGLMIGVELVEPGTKKPLPVSLVKDIHEEIKDNGVLVVLGGRWSNVFRMKPPMCITKADVDFEKHIEMANRGTKLCFDIVRTYSTAKMPPTDFVPRPYTGPSYQQVEQMKGVYMPPSITNAYKKPVLLTQGHMQWLYDNDGKRYLDLFGGIVTVSVGHCHPKVNAALKDQLDVLWHTTNLYRHPKIYEYVEQLAAKLPGDLNVVYLVNSGSEANELATLLAKAYTGNLDIISLQTSYHGYTSSLMGLTATQSYRMAIPVPPGFYHAVHPDPFRGAFGGCRDSISQAPGSCSCTGECISTDKYVHQLNELLGNSVPAGRVAAFFAESIQGAGGVVQFPKGYLKKAQELIKKNGGLYVADEVQTGFGRTGDHFWGFETHGVKPDIVTMAKGIGNGFPLAAVVTTKEIAANHAKAAYFNTFGGNPMASTVGKAVLEVIEEEGLQQNSKVVGEYFIRQLMDLQKQHPVIGDVRGQGLMIGVELVEPGTKTPLTTSKVNDIHENIKDNGVLIARGGRFNNVFRIKPPMCITKQDVDFGISIINDAIKKVVN
Summary
Uniprot
Pubmed
EMBL
Proteomes
Pfam
PF00202 Aminotran_3
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
CDD
ProteinModelPortal
PDB
5G4J
E-value=7.67724e-59,
Score=579
Ontologies
PATHWAY
00250
Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00260 Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00260 Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
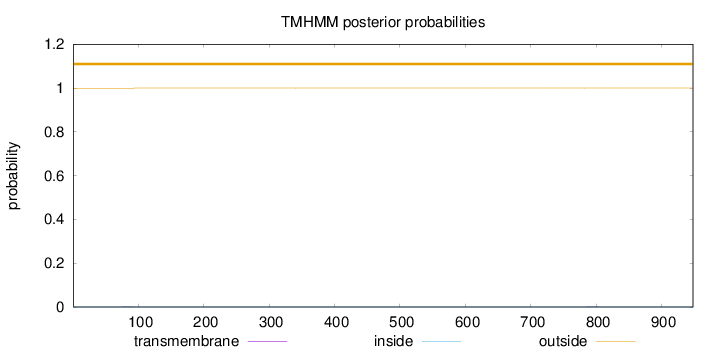
Topology
Length:
948
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0170099999999999
Exp number, first 60 AAs:
0.00137
Total prob of N-in:
0.00056
outside
1 - 948
Population Genetic Test Statistics
Pi
213.689639
Theta
184.930447
Tajima's D
1.091691
CLR
0.260541
CSRT
0.686965651717414
Interpretation
Uncertain
