Gene
KWMTBOMO06427 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011599
Annotation
signal_sequence_receptor_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.018
Sequence
CDS
ATGAAATTAATATATTTTACGTTCTTGATTGTACCGGCGGCTTTATTATGCCTTGGAAGCGGTAACATCTTAGCTAGAGCAGAAGAGGATGAACTTGATGATGTTGTCGACATTGAAGGAGAAGACAACCAGGTAGTCGGTGACGATGCTCTCGTAGATGATGATGAGAGTGTTGTTAAGTCATCTCAGGATATTGATACAACAATACTGTTCACCAAACCTGTGCCTAGCTTGGGTGACTTAACTTTCGATATTCAAGCTGGTTACCCAGTGGAATTTTTTGTGGGCTTCATCAACAAGGGTTCCGTAGACTATGTAGTAGAATCCATGGAGGCATCTTTCCGGTATCCAATGGACTACACATACTACATTCAGAACTTCACCGCGTTGCCTTATAACAGAGAAGTTAAGCCAAAACAGGAAGCCACCTTTGCCTATTCCTTCATCCCCAATGAAGCTTTTGCTGGAAGACCTTTCGGATTAAATGTGCAGCTTAATTACAGAGATGCCAGCGGAAATGCTTACCAGGAAGCAGTCTACAACCAGACACTCAACATTGTGGAAGTATCCGAAGGTCTAGATGGAGAGACATTCTTCTTATATGTCTTCCTTGGTGCTGGTGGAGTGTTGGCTCTCGTCGCAGGACAGCAAGCCCTCGCTTCATTGGCCAGACGTAGATCACCACGTTCAGTGTCTCAGCCCGTTGAAACTGGAACAGCCAGTGAAGTAGACTATGACTGGCTACCTAAAGAAATTGTGAATCAACTCAAAAAATCTCCCAAGACGCCGAAGCAGTCGCCGCGCGCAAGGAAAGCAAAGAGATCAGCAGGCGACGACTAA
Protein
MKLIYFTFLIVPAALLCLGSGNILARAEEDELDDVVDIEGEDNQVVGDDALVDDDESVVKSSQDIDTTILFTKPVPSLGDLTFDIQAGYPVEFFVGFINKGSVDYVVESMEASFRYPMDYTYYIQNFTALPYNREVKPKQEATFAYSFIPNEAFAGRPFGLNVQLNYRDASGNAYQEAVYNQTLNIVEVSEGLDGETFFLYVFLGAGGVLALVAGQQALASLARRRSPRSVSQPVETGTASEVDYDWLPKEIVNQLKKSPKTPKQSPRARKAKRSAGDD
Summary
Uniprot
Q2F5L0
A0A3G1T1J9
A0A3S2L7F1
I4DJ05
A0A0N1PH20
I4DN16
+ More
S4PBB9 A0A1E1WT20 A0A2W1BUL3 A0A2A4J1I6 A0A2H1W4W5 E3UKP9 A0A212FK41 A0A2J7PCZ7 A0A023ENQ8 A0A182GFZ8 A0A023EMK7 A0A232F8U9 A0A0P6IT12 A0A170Y3V2 B0XFI4 U5ESU1 R4FQ38 B8RJF4 A0A0C9QRG9 A0A1Q3FEI3 A0A067REZ9 A0A224XUJ4 A0A182VEJ6 A0A182U753 A0A182LA29 Q7PY28 A0A182HLP0 A0A182WST6 A0A026WL01 D6WHE5 A0A182RZN0 A0A224XTP2 A0A182JU80 A0A1L8DUH2 A0A182PSI5 A0A0T6B8A2 A0A182MBG6 A0A1B6M708 A0A182VZM4 A0A0L7QNT4 A0A1J1IA74 A0A182NRT4 A0A182Y3T4 A0A2M4A713 T1E8B2 A0A2M3YZZ7 A0A158NXZ7 A0A2A3EF06 A0A0N0U384 W5JBF1 A0A2M4A577 A0A088A6M3 B3N070 A0A2M4BVW9 A0A182FPJ3 Q9W335 B3NT65 A0A182QGY5 A0A336LU05 A0A182J4Z6 B4PWX0 B4IDH0 A0A336L2A4 A0A1Y1KFE3 A0A084VLC1 A0A0P4WHD0 A0A1W4XUL7 B4JIP8 A0A1B6CHQ9 A0A154P1F4 B4NDZ9 A0A1L8EHJ8 A0A195ED21 A0A2P8XM45 W8BVK4 A0A034WKP4 E2B9X6 A0A1I8PMP3 A0A0A1WJG2 B4M220 B4L8F1 A0A146LEQ3 A0A0A9YTV1 A0A170Y3R8 A0A0K8UBB9 B4GVB2 Q29ID3 J3JZ13 T1DJS8 A0A1W4VHS9 A0A3B0KNX4 A0A0M4EZB7 R4WDU7
S4PBB9 A0A1E1WT20 A0A2W1BUL3 A0A2A4J1I6 A0A2H1W4W5 E3UKP9 A0A212FK41 A0A2J7PCZ7 A0A023ENQ8 A0A182GFZ8 A0A023EMK7 A0A232F8U9 A0A0P6IT12 A0A170Y3V2 B0XFI4 U5ESU1 R4FQ38 B8RJF4 A0A0C9QRG9 A0A1Q3FEI3 A0A067REZ9 A0A224XUJ4 A0A182VEJ6 A0A182U753 A0A182LA29 Q7PY28 A0A182HLP0 A0A182WST6 A0A026WL01 D6WHE5 A0A182RZN0 A0A224XTP2 A0A182JU80 A0A1L8DUH2 A0A182PSI5 A0A0T6B8A2 A0A182MBG6 A0A1B6M708 A0A182VZM4 A0A0L7QNT4 A0A1J1IA74 A0A182NRT4 A0A182Y3T4 A0A2M4A713 T1E8B2 A0A2M3YZZ7 A0A158NXZ7 A0A2A3EF06 A0A0N0U384 W5JBF1 A0A2M4A577 A0A088A6M3 B3N070 A0A2M4BVW9 A0A182FPJ3 Q9W335 B3NT65 A0A182QGY5 A0A336LU05 A0A182J4Z6 B4PWX0 B4IDH0 A0A336L2A4 A0A1Y1KFE3 A0A084VLC1 A0A0P4WHD0 A0A1W4XUL7 B4JIP8 A0A1B6CHQ9 A0A154P1F4 B4NDZ9 A0A1L8EHJ8 A0A195ED21 A0A2P8XM45 W8BVK4 A0A034WKP4 E2B9X6 A0A1I8PMP3 A0A0A1WJG2 B4M220 B4L8F1 A0A146LEQ3 A0A0A9YTV1 A0A170Y3R8 A0A0K8UBB9 B4GVB2 Q29ID3 J3JZ13 T1DJS8 A0A1W4VHS9 A0A3B0KNX4 A0A0M4EZB7 R4WDU7
Pubmed
19121390
22651552
26354079
23622113
28756777
22118469
+ More
24945155 26483478 28648823 26999592 24845553 20966253 12364791 14747013 15944077 17210077 24508170 30249741 18362917 19820115 25244985 21347285 20920257 23761445 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28004739 24438588 29403074 24495485 25348373 20798317 25830018 26823975 25401762 15632085 22516182 24330624 23691247
24945155 26483478 28648823 26999592 24845553 20966253 12364791 14747013 15944077 17210077 24508170 30249741 18362917 19820115 25244985 21347285 20920257 23761445 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28004739 24438588 29403074 24495485 25348373 20798317 25830018 26823975 25401762 15632085 22516182 24330624 23691247
EMBL
BABH01012407
BABH01012408
DQ311413
ABD36357.1
MG992404
AXY94842.1
+ More
RSAL01000104 RVE47397.1 AK401273 KQ459590 BAM17895.1 KPI97173.1 KQ460971 KPJ10140.1 AK402684 BAM19306.1 GAIX01005987 JAA86573.1 GDQN01000879 JAT90175.1 KZ149896 PZC78738.1 NWSH01003679 PCG65945.1 ODYU01006349 SOQ48121.1 HM449923 ADO33058.1 AGBW02008141 OWR54113.1 NEVH01026401 PNF14198.1 GAPW01003027 JAC10571.1 JXUM01060986 KQ562129 KXJ76626.1 GAPW01003026 JAC10572.1 NNAY01000704 OXU26889.1 GDUN01000987 JAN94932.1 GEMB01003693 JAR99527.1 DS232930 EDS26822.1 GANO01003095 JAB56776.1 ACPB03016596 GAHY01000609 JAA76901.1 EZ000579 ACJ64453.1 GBYB01006284 JAG76051.1 GFDL01009078 JAV25967.1 KK852729 KDR17543.1 GFTR01004805 JAW11621.1 CR954257 AAAB01008987 CAJ14158.1 EAA01377.3 APCN01000909 KK107167 QOIP01000010 EZA56351.1 RLU17850.1 KQ971321 EFA00653.1 GFTR01004953 JAW11473.1 GFDF01004000 JAV10084.1 LJIG01009323 KRT83309.1 AXCM01000026 GEBQ01008296 JAT31681.1 KQ414843 KOC60302.1 CVRI01000044 CRK96468.1 GGFK01003266 MBW36587.1 GAMD01002923 JAA98667.1 GGFM01001065 MBW21816.1 ADTU01002828 ADTU01002829 KZ288263 PBC30300.1 KQ435955 KOX68044.1 ADMH02001712 ETN61326.1 GGFK01002645 MBW35966.1 CH902640 EDV38274.1 GGFJ01008068 MBW57209.1 AE014298 AY069541 AAF46502.1 AAL39686.1 AHN59528.1 CH954180 EDV46245.1 AXCN02001148 UFQT01000110 SSX20163.1 CM000162 EDX02854.1 CH480830 EDW45628.1 UFQS01001536 UFQT01001536 SSX11396.1 SSX30964.1 GEZM01087017 JAV58910.1 ATLV01014444 KE524972 KFB38765.1 GDRN01053456 GDRN01053455 JAI66170.1 CH916370 EDW00495.1 GEDC01024319 JAS12979.1 KQ434786 KZC05174.1 CH964239 EDW81968.1 GFDG01000674 JAV18125.1 KQ979074 KYN22717.1 PYGN01001739 PSN33071.1 GAMC01005612 JAC00944.1 GAKP01002801 JAC56151.1 GL446605 EFN87563.1 GBXI01015300 JAC98991.1 CH940651 EDW65724.1 CH933815 EDW07926.1 GDHC01013619 JAQ05010.1 GBHO01008558 JAG35046.1 GEMB01003694 JAR99526.1 GDHF01028441 JAI23873.1 CH479192 EDW26649.1 CH379063 EAL32720.1 BT128494 AEE63451.1 GALA01000081 JAA94771.1 OUUW01000011 SPP86851.1 CP012528 ALC49492.1 AK417864 BAN21079.1
RSAL01000104 RVE47397.1 AK401273 KQ459590 BAM17895.1 KPI97173.1 KQ460971 KPJ10140.1 AK402684 BAM19306.1 GAIX01005987 JAA86573.1 GDQN01000879 JAT90175.1 KZ149896 PZC78738.1 NWSH01003679 PCG65945.1 ODYU01006349 SOQ48121.1 HM449923 ADO33058.1 AGBW02008141 OWR54113.1 NEVH01026401 PNF14198.1 GAPW01003027 JAC10571.1 JXUM01060986 KQ562129 KXJ76626.1 GAPW01003026 JAC10572.1 NNAY01000704 OXU26889.1 GDUN01000987 JAN94932.1 GEMB01003693 JAR99527.1 DS232930 EDS26822.1 GANO01003095 JAB56776.1 ACPB03016596 GAHY01000609 JAA76901.1 EZ000579 ACJ64453.1 GBYB01006284 JAG76051.1 GFDL01009078 JAV25967.1 KK852729 KDR17543.1 GFTR01004805 JAW11621.1 CR954257 AAAB01008987 CAJ14158.1 EAA01377.3 APCN01000909 KK107167 QOIP01000010 EZA56351.1 RLU17850.1 KQ971321 EFA00653.1 GFTR01004953 JAW11473.1 GFDF01004000 JAV10084.1 LJIG01009323 KRT83309.1 AXCM01000026 GEBQ01008296 JAT31681.1 KQ414843 KOC60302.1 CVRI01000044 CRK96468.1 GGFK01003266 MBW36587.1 GAMD01002923 JAA98667.1 GGFM01001065 MBW21816.1 ADTU01002828 ADTU01002829 KZ288263 PBC30300.1 KQ435955 KOX68044.1 ADMH02001712 ETN61326.1 GGFK01002645 MBW35966.1 CH902640 EDV38274.1 GGFJ01008068 MBW57209.1 AE014298 AY069541 AAF46502.1 AAL39686.1 AHN59528.1 CH954180 EDV46245.1 AXCN02001148 UFQT01000110 SSX20163.1 CM000162 EDX02854.1 CH480830 EDW45628.1 UFQS01001536 UFQT01001536 SSX11396.1 SSX30964.1 GEZM01087017 JAV58910.1 ATLV01014444 KE524972 KFB38765.1 GDRN01053456 GDRN01053455 JAI66170.1 CH916370 EDW00495.1 GEDC01024319 JAS12979.1 KQ434786 KZC05174.1 CH964239 EDW81968.1 GFDG01000674 JAV18125.1 KQ979074 KYN22717.1 PYGN01001739 PSN33071.1 GAMC01005612 JAC00944.1 GAKP01002801 JAC56151.1 GL446605 EFN87563.1 GBXI01015300 JAC98991.1 CH940651 EDW65724.1 CH933815 EDW07926.1 GDHC01013619 JAQ05010.1 GBHO01008558 JAG35046.1 GEMB01003694 JAR99526.1 GDHF01028441 JAI23873.1 CH479192 EDW26649.1 CH379063 EAL32720.1 BT128494 AEE63451.1 GALA01000081 JAA94771.1 OUUW01000011 SPP86851.1 CP012528 ALC49492.1 AK417864 BAN21079.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000218220
UP000007151
+ More
UP000235965 UP000069940 UP000249989 UP000215335 UP000002320 UP000015103 UP000027135 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000076407 UP000053097 UP000279307 UP000007266 UP000075900 UP000075881 UP000075885 UP000075883 UP000075920 UP000053825 UP000183832 UP000075884 UP000076408 UP000005205 UP000242457 UP000053105 UP000000673 UP000005203 UP000007801 UP000069272 UP000000803 UP000008711 UP000075886 UP000075880 UP000002282 UP000001292 UP000030765 UP000192223 UP000001070 UP000076502 UP000007798 UP000078492 UP000245037 UP000008237 UP000095300 UP000008792 UP000009192 UP000008744 UP000001819 UP000192221 UP000268350 UP000092553
UP000235965 UP000069940 UP000249989 UP000215335 UP000002320 UP000015103 UP000027135 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000076407 UP000053097 UP000279307 UP000007266 UP000075900 UP000075881 UP000075885 UP000075883 UP000075920 UP000053825 UP000183832 UP000075884 UP000076408 UP000005205 UP000242457 UP000053105 UP000000673 UP000005203 UP000007801 UP000069272 UP000000803 UP000008711 UP000075886 UP000075880 UP000002282 UP000001292 UP000030765 UP000192223 UP000001070 UP000076502 UP000007798 UP000078492 UP000245037 UP000008237 UP000095300 UP000008792 UP000009192 UP000008744 UP000001819 UP000192221 UP000268350 UP000092553
Interpro
SUPFAM
SSF53254
SSF53254
Gene 3D
ProteinModelPortal
Q2F5L0
A0A3G1T1J9
A0A3S2L7F1
I4DJ05
A0A0N1PH20
I4DN16
+ More
S4PBB9 A0A1E1WT20 A0A2W1BUL3 A0A2A4J1I6 A0A2H1W4W5 E3UKP9 A0A212FK41 A0A2J7PCZ7 A0A023ENQ8 A0A182GFZ8 A0A023EMK7 A0A232F8U9 A0A0P6IT12 A0A170Y3V2 B0XFI4 U5ESU1 R4FQ38 B8RJF4 A0A0C9QRG9 A0A1Q3FEI3 A0A067REZ9 A0A224XUJ4 A0A182VEJ6 A0A182U753 A0A182LA29 Q7PY28 A0A182HLP0 A0A182WST6 A0A026WL01 D6WHE5 A0A182RZN0 A0A224XTP2 A0A182JU80 A0A1L8DUH2 A0A182PSI5 A0A0T6B8A2 A0A182MBG6 A0A1B6M708 A0A182VZM4 A0A0L7QNT4 A0A1J1IA74 A0A182NRT4 A0A182Y3T4 A0A2M4A713 T1E8B2 A0A2M3YZZ7 A0A158NXZ7 A0A2A3EF06 A0A0N0U384 W5JBF1 A0A2M4A577 A0A088A6M3 B3N070 A0A2M4BVW9 A0A182FPJ3 Q9W335 B3NT65 A0A182QGY5 A0A336LU05 A0A182J4Z6 B4PWX0 B4IDH0 A0A336L2A4 A0A1Y1KFE3 A0A084VLC1 A0A0P4WHD0 A0A1W4XUL7 B4JIP8 A0A1B6CHQ9 A0A154P1F4 B4NDZ9 A0A1L8EHJ8 A0A195ED21 A0A2P8XM45 W8BVK4 A0A034WKP4 E2B9X6 A0A1I8PMP3 A0A0A1WJG2 B4M220 B4L8F1 A0A146LEQ3 A0A0A9YTV1 A0A170Y3R8 A0A0K8UBB9 B4GVB2 Q29ID3 J3JZ13 T1DJS8 A0A1W4VHS9 A0A3B0KNX4 A0A0M4EZB7 R4WDU7
S4PBB9 A0A1E1WT20 A0A2W1BUL3 A0A2A4J1I6 A0A2H1W4W5 E3UKP9 A0A212FK41 A0A2J7PCZ7 A0A023ENQ8 A0A182GFZ8 A0A023EMK7 A0A232F8U9 A0A0P6IT12 A0A170Y3V2 B0XFI4 U5ESU1 R4FQ38 B8RJF4 A0A0C9QRG9 A0A1Q3FEI3 A0A067REZ9 A0A224XUJ4 A0A182VEJ6 A0A182U753 A0A182LA29 Q7PY28 A0A182HLP0 A0A182WST6 A0A026WL01 D6WHE5 A0A182RZN0 A0A224XTP2 A0A182JU80 A0A1L8DUH2 A0A182PSI5 A0A0T6B8A2 A0A182MBG6 A0A1B6M708 A0A182VZM4 A0A0L7QNT4 A0A1J1IA74 A0A182NRT4 A0A182Y3T4 A0A2M4A713 T1E8B2 A0A2M3YZZ7 A0A158NXZ7 A0A2A3EF06 A0A0N0U384 W5JBF1 A0A2M4A577 A0A088A6M3 B3N070 A0A2M4BVW9 A0A182FPJ3 Q9W335 B3NT65 A0A182QGY5 A0A336LU05 A0A182J4Z6 B4PWX0 B4IDH0 A0A336L2A4 A0A1Y1KFE3 A0A084VLC1 A0A0P4WHD0 A0A1W4XUL7 B4JIP8 A0A1B6CHQ9 A0A154P1F4 B4NDZ9 A0A1L8EHJ8 A0A195ED21 A0A2P8XM45 W8BVK4 A0A034WKP4 E2B9X6 A0A1I8PMP3 A0A0A1WJG2 B4M220 B4L8F1 A0A146LEQ3 A0A0A9YTV1 A0A170Y3R8 A0A0K8UBB9 B4GVB2 Q29ID3 J3JZ13 T1DJS8 A0A1W4VHS9 A0A3B0KNX4 A0A0M4EZB7 R4WDU7
Ontologies
GO
Topology
SignalP
Position: 1 - 27,
Likelihood: 0.948528
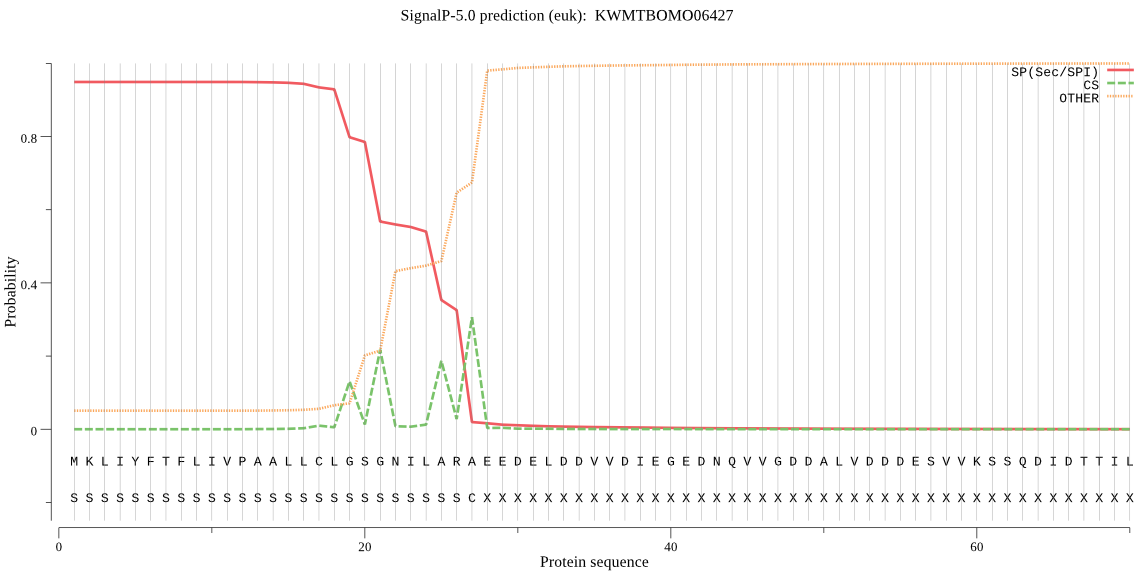
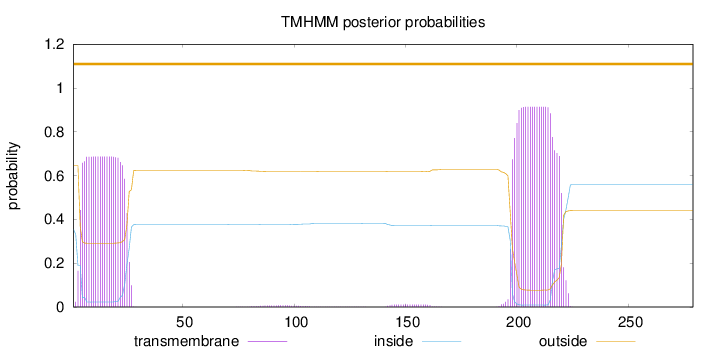
Length:
279
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
35.30629
Exp number, first 60 AAs:
14.87186
Total prob of N-in:
0.35513
POSSIBLE N-term signal
sequence
outside
1 - 279
Population Genetic Test Statistics
Pi
277.072247
Theta
212.289654
Tajima's D
0.941585
CLR
1.302918
CSRT
0.643617819109045
Interpretation
Uncertain
