Gene
KWMTBOMO06426 Validated by peptides from experiments
Annotation
PREDICTED:_alanine--glyoxylate_aminotransferase_2?_mitochondrial_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.582 Mitochondrial Reliability : 1.186
Sequence
CDS
ATGATGGCATTGACTGCAATGCAAAGCTTCAGGCAACCAATACCAGTTCCCCCTGGATTTCATCACGCCATGGTTCCAGATCCATTTAGAAATGAATGGGGTGGATGTCGGGATTGCATTTCTCAAGTGCCAGACTCGTGCTCCTGTACGGGAGATTGTCTTTCTACTGATAAATATATCGATCAATTGAAAGAGGTTCTTGAGTTTTCGATACCTAGTCGACGGGTAGCGGCGTTCTTTGCTGAGTCTATCCAGGGGGCTGGAGGGATCGTGCAGTTCCCAAAGGGTTACATTAAGAGAGCTCAACAGCTTATTAAAGAGCATGGTGGCATATTTGTTGCTGACGAGGTCCAGACTGGTTTTGCGCGTACTGGTGACACTTTCTGGGGTTTCGAATCACACGGTATCATTCCTGACATAGTAACGCTGGCGAAAGGCATCGCTAACGGGTTGCCGTTTGCAGCTGTAGTTACGACCAAAGAAATCGCCGAGGCACATACTAAAGCGAATTACTTTAACACTTTTAGTGGCAATGCTCTGGCGGCAGCAGCTGGCGTGGCTGTATTGGAGGTGATTGACGATGAAAACATTCAAGATCATTGCAAGAAGATAGGAGAATATTTTATTCGTCAATTAATACAGCTTCAACAACAATTTCCGATTATTGGTGACGTCAGGGGTAAAGGTCTGATGTTGGCAATGGAACTAGTTAGACCAGGCACTAAAGAAAATTTGAGTGACAATTGCATGATAAGAATGCATCAAAGATTAAAAGATCTTGGCGTTTTGACTGGTCTTGGTGGTCGGAACCTAAATGTCTTGAAAATAACTCCACCGATGTGTATTACTAAAGAAGACGTGGATTTTGCAGTATCCGTTTTTAATGATGTTTTTAAGCAATTCTCTTAA
Protein
MMALTAMQSFRQPIPVPPGFHHAMVPDPFRNEWGGCRDCISQVPDSCSCTGDCLSTDKYIDQLKEVLEFSIPSRRVAAFFAESIQGAGGIVQFPKGYIKRAQQLIKEHGGIFVADEVQTGFARTGDTFWGFESHGIIPDIVTLAKGIANGLPFAAVVTTKEIAEAHTKANYFNTFSGNALAAAAGVAVLEVIDDENIQDHCKKIGEYFIRQLIQLQQQFPIIGDVRGKGLMLAMELVRPGTKENLSDNCMIRMHQRLKDLGVLTGLGGRNLNVLKITPPMCITKEDVDFAVSVFNDVFKQFS
Summary
Similarity
Belongs to the class-III pyridoxal-phosphate-dependent aminotransferase family.
Uniprot
H9JPZ5
A0A212FK51
A0A0N0PBB4
A0A2W1C0V7
A0A3S2NH35
A0A194PWN7
+ More
A0A0N1I7H1 A0A2H1VJE8 H9JPZ6 A0A226DS77 U5EHG1 A0A1Q3FIX3 Q17NN1 T1IEJ9 T1DLB3 A0A1Y9IUA8 A0A2R7VZ99 A0A023F4P8 A0A224XBX3 A0A0P4VHK7 A0A0L8FK67 A0A1D1VKF4 A0A1Y1KAE8 A0A1Q3FIG9 A0A2M4BL18 A0A2M4BLV8 A0A0K8THH4 A0A0K2TFI8 A0A1I8MSC5 A0A182WZT6 Q5TMY1 A0A182WEX6 A0A182JE15 A0A2C9GKX9 T1P817 T1PKC2 A0A1Q3FID0 A0A0K8U5Q3 A0A0K8WKD6 A0A1B6GM39 A0A1B6KHZ6 B0X3N0 B0WFQ7 A0A084WIX2 A0A1V9XS66 A0A182Y1G8 A0A182QW57 A0A2M4CYM1 Q17NN2 A0A182MYT1 A0A023GNL2 A0A1E1XFD1 A0A0A1WWK2 A0A210QH45 A0A2M4ASF1 A0A0A1WN51 K1RB89 A0A182V1D5 A0A2J7QLT3 A0A1J1IBT4 A0A0L0C895 W5JIX0 A0A336M3H9 A0A182HQD2 A0A0R1DZM6 A0A1B6M7X2 A0A336LW49 A0A0A1WFV2 B4N5G0 A0A182P5V5 L7M8G8 B3NEF3 A0A131XYN6 A0A336LR78 A8E6R2 A0A1I8Q0P5 W8B5N7 V5GUJ9 A0A1S3IEV7 Q9VNR7 Q95TT3 A0A336LRV8 A0A0B7AD12 A0A293MBL7 A0A1A9VFA7 E9GEQ7 A0A224YT48 A0A1B0A2D4 B4PIP8 A0A131Z5C2 T1KR84 B3MB82 G3MK55 T1JJ22 Q29F13 H3A3S3 A0A1W4UR20 B4LDY2 A0A0C9RF91 A0A0J9RZY9
A0A0N1I7H1 A0A2H1VJE8 H9JPZ6 A0A226DS77 U5EHG1 A0A1Q3FIX3 Q17NN1 T1IEJ9 T1DLB3 A0A1Y9IUA8 A0A2R7VZ99 A0A023F4P8 A0A224XBX3 A0A0P4VHK7 A0A0L8FK67 A0A1D1VKF4 A0A1Y1KAE8 A0A1Q3FIG9 A0A2M4BL18 A0A2M4BLV8 A0A0K8THH4 A0A0K2TFI8 A0A1I8MSC5 A0A182WZT6 Q5TMY1 A0A182WEX6 A0A182JE15 A0A2C9GKX9 T1P817 T1PKC2 A0A1Q3FID0 A0A0K8U5Q3 A0A0K8WKD6 A0A1B6GM39 A0A1B6KHZ6 B0X3N0 B0WFQ7 A0A084WIX2 A0A1V9XS66 A0A182Y1G8 A0A182QW57 A0A2M4CYM1 Q17NN2 A0A182MYT1 A0A023GNL2 A0A1E1XFD1 A0A0A1WWK2 A0A210QH45 A0A2M4ASF1 A0A0A1WN51 K1RB89 A0A182V1D5 A0A2J7QLT3 A0A1J1IBT4 A0A0L0C895 W5JIX0 A0A336M3H9 A0A182HQD2 A0A0R1DZM6 A0A1B6M7X2 A0A336LW49 A0A0A1WFV2 B4N5G0 A0A182P5V5 L7M8G8 B3NEF3 A0A131XYN6 A0A336LR78 A8E6R2 A0A1I8Q0P5 W8B5N7 V5GUJ9 A0A1S3IEV7 Q9VNR7 Q95TT3 A0A336LRV8 A0A0B7AD12 A0A293MBL7 A0A1A9VFA7 E9GEQ7 A0A224YT48 A0A1B0A2D4 B4PIP8 A0A131Z5C2 T1KR84 B3MB82 G3MK55 T1JJ22 Q29F13 H3A3S3 A0A1W4UR20 B4LDY2 A0A0C9RF91 A0A0J9RZY9
Pubmed
19121390
22118469
26354079
28756777
17510324
25474469
+ More
27129103 27649274 28004739 26823975 25315136 12364791 14747013 17210077 24438588 28327890 25244985 28503490 25830018 28812685 22992520 26108605 20920257 23761445 17994087 17550304 25576852 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 25765539 21292972 28797301 26830274 22216098 15632085 9215903 22936249
27129103 27649274 28004739 26823975 25315136 12364791 14747013 17210077 24438588 28327890 25244985 28503490 25830018 28812685 22992520 26108605 20920257 23761445 17994087 17550304 25576852 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 25765539 21292972 28797301 26830274 22216098 15632085 9215903 22936249
EMBL
BABH01012406
AGBW02008141
OWR54114.1
KQ460971
KPJ10138.1
KZ149896
+ More
PZC78740.1 RSAL01000104 RVE47398.1 KQ459590 KPI97174.1 KPJ10139.1 ODYU01002894 SOQ40953.1 BABH01012403 BABH01012404 BABH01012405 LNIX01000014 OXA47066.1 GANO01003035 JAB56836.1 GFDL01007485 JAV27560.1 CH477198 EAT48281.1 ACPB03014069 GAMD01000541 JAB01050.1 KK854169 PTY12508.1 GBBI01002763 JAC15949.1 GFTR01006539 JAW09887.1 GDKW01002596 JAI53999.1 KQ429994 KOF64825.1 BDGG01000008 GAV02097.1 GEZM01087795 JAV58472.1 GFDL01007644 JAV27401.1 GGFJ01004628 MBW53769.1 GGFJ01004627 MBW53768.1 GBRD01001221 GDHC01020519 GDHC01008915 JAG64600.1 JAP98109.1 JAQ09714.1 HACA01006865 CDW24226.1 AAAB01008986 EAL38668.3 APCN01003309 KA644689 AFP59318.1 KA649211 AFP63840.1 GFDL01007704 JAV27341.1 GDHF01030340 JAI21974.1 GDHF01000762 JAI51552.1 GECZ01006386 JAS63383.1 GEBQ01028908 JAT11069.1 DS232318 EDS39923.1 DS231920 EDS26402.1 ATLV01023952 KE525347 KFB50166.1 MNPL01004920 OQR76350.1 AXCN02001025 GGFL01006255 MBW70433.1 EAT48282.1 GBBM01000938 JAC34480.1 GFAC01001256 JAT97932.1 GBXI01011266 JAD03026.1 NEDP02003738 OWF48054.1 GGFK01010341 MBW43662.1 GBXI01014362 JAC99929.1 JH815890 EKC31351.1 NEVH01013243 PNF29508.1 CVRI01000047 CRK97743.1 JRES01000763 KNC28486.1 ADMH02001279 ETN63238.1 UFQT01000127 SSX20548.1 CM000159 KRK02512.1 GEBQ01020729 GEBQ01007951 JAT19248.1 JAT32026.1 SSX20547.1 GBXI01017014 JAC97277.1 CH964101 EDW79599.1 GACK01004887 JAA60147.1 CH954178 EDV52788.2 GEFM01004970 JAP70826.1 SSX20546.1 AE014296 BT030854 AAN12201.2 ABV82236.1 ALI30495.1 GAMC01021467 GAMC01021466 GAMC01021465 GAMC01021464 JAB85089.1 GANP01010403 JAB74065.1 BT044304 AAF51855.3 ACH92369.1 AY058552 AAL13781.1 SSX20545.1 HACG01030985 CEK77850.1 GFWV01012742 MAA37471.1 GL732541 EFX81878.1 GFPF01005854 MAA17000.1 EDW95556.2 GEDV01001983 JAP86574.1 CAEY01000384 CH902618 EDV41383.2 JO842256 AEO33873.1 JH431841 CH379070 EAL29896.2 AFYH01129082 AFYH01129083 AFYH01129084 AFYH01129085 AFYH01129086 AFYH01129087 AFYH01129088 AFYH01129089 CH940647 EDW70025.2 GBYB01007080 JAG76847.1 CM002912 KMZ01222.1
PZC78740.1 RSAL01000104 RVE47398.1 KQ459590 KPI97174.1 KPJ10139.1 ODYU01002894 SOQ40953.1 BABH01012403 BABH01012404 BABH01012405 LNIX01000014 OXA47066.1 GANO01003035 JAB56836.1 GFDL01007485 JAV27560.1 CH477198 EAT48281.1 ACPB03014069 GAMD01000541 JAB01050.1 KK854169 PTY12508.1 GBBI01002763 JAC15949.1 GFTR01006539 JAW09887.1 GDKW01002596 JAI53999.1 KQ429994 KOF64825.1 BDGG01000008 GAV02097.1 GEZM01087795 JAV58472.1 GFDL01007644 JAV27401.1 GGFJ01004628 MBW53769.1 GGFJ01004627 MBW53768.1 GBRD01001221 GDHC01020519 GDHC01008915 JAG64600.1 JAP98109.1 JAQ09714.1 HACA01006865 CDW24226.1 AAAB01008986 EAL38668.3 APCN01003309 KA644689 AFP59318.1 KA649211 AFP63840.1 GFDL01007704 JAV27341.1 GDHF01030340 JAI21974.1 GDHF01000762 JAI51552.1 GECZ01006386 JAS63383.1 GEBQ01028908 JAT11069.1 DS232318 EDS39923.1 DS231920 EDS26402.1 ATLV01023952 KE525347 KFB50166.1 MNPL01004920 OQR76350.1 AXCN02001025 GGFL01006255 MBW70433.1 EAT48282.1 GBBM01000938 JAC34480.1 GFAC01001256 JAT97932.1 GBXI01011266 JAD03026.1 NEDP02003738 OWF48054.1 GGFK01010341 MBW43662.1 GBXI01014362 JAC99929.1 JH815890 EKC31351.1 NEVH01013243 PNF29508.1 CVRI01000047 CRK97743.1 JRES01000763 KNC28486.1 ADMH02001279 ETN63238.1 UFQT01000127 SSX20548.1 CM000159 KRK02512.1 GEBQ01020729 GEBQ01007951 JAT19248.1 JAT32026.1 SSX20547.1 GBXI01017014 JAC97277.1 CH964101 EDW79599.1 GACK01004887 JAA60147.1 CH954178 EDV52788.2 GEFM01004970 JAP70826.1 SSX20546.1 AE014296 BT030854 AAN12201.2 ABV82236.1 ALI30495.1 GAMC01021467 GAMC01021466 GAMC01021465 GAMC01021464 JAB85089.1 GANP01010403 JAB74065.1 BT044304 AAF51855.3 ACH92369.1 AY058552 AAL13781.1 SSX20545.1 HACG01030985 CEK77850.1 GFWV01012742 MAA37471.1 GL732541 EFX81878.1 GFPF01005854 MAA17000.1 EDW95556.2 GEDV01001983 JAP86574.1 CAEY01000384 CH902618 EDV41383.2 JO842256 AEO33873.1 JH431841 CH379070 EAL29896.2 AFYH01129082 AFYH01129083 AFYH01129084 AFYH01129085 AFYH01129086 AFYH01129087 AFYH01129088 AFYH01129089 CH940647 EDW70025.2 GBYB01007080 JAG76847.1 CM002912 KMZ01222.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000283053
UP000053268
UP000198287
+ More
UP000008820 UP000015103 UP000075920 UP000053454 UP000186922 UP000095301 UP000076407 UP000007062 UP000075880 UP000075840 UP000002320 UP000030765 UP000192247 UP000076408 UP000075886 UP000075884 UP000242188 UP000005408 UP000075903 UP000235965 UP000183832 UP000037069 UP000000673 UP000002282 UP000007798 UP000075885 UP000008711 UP000000803 UP000095300 UP000085678 UP000078200 UP000000305 UP000092445 UP000015104 UP000007801 UP000001819 UP000008672 UP000192221 UP000008792
UP000008820 UP000015103 UP000075920 UP000053454 UP000186922 UP000095301 UP000076407 UP000007062 UP000075880 UP000075840 UP000002320 UP000030765 UP000192247 UP000076408 UP000075886 UP000075884 UP000242188 UP000005408 UP000075903 UP000235965 UP000183832 UP000037069 UP000000673 UP000002282 UP000007798 UP000075885 UP000008711 UP000000803 UP000095300 UP000085678 UP000078200 UP000000305 UP000092445 UP000015104 UP000007801 UP000001819 UP000008672 UP000192221 UP000008792
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JPZ5
A0A212FK51
A0A0N0PBB4
A0A2W1C0V7
A0A3S2NH35
A0A194PWN7
+ More
A0A0N1I7H1 A0A2H1VJE8 H9JPZ6 A0A226DS77 U5EHG1 A0A1Q3FIX3 Q17NN1 T1IEJ9 T1DLB3 A0A1Y9IUA8 A0A2R7VZ99 A0A023F4P8 A0A224XBX3 A0A0P4VHK7 A0A0L8FK67 A0A1D1VKF4 A0A1Y1KAE8 A0A1Q3FIG9 A0A2M4BL18 A0A2M4BLV8 A0A0K8THH4 A0A0K2TFI8 A0A1I8MSC5 A0A182WZT6 Q5TMY1 A0A182WEX6 A0A182JE15 A0A2C9GKX9 T1P817 T1PKC2 A0A1Q3FID0 A0A0K8U5Q3 A0A0K8WKD6 A0A1B6GM39 A0A1B6KHZ6 B0X3N0 B0WFQ7 A0A084WIX2 A0A1V9XS66 A0A182Y1G8 A0A182QW57 A0A2M4CYM1 Q17NN2 A0A182MYT1 A0A023GNL2 A0A1E1XFD1 A0A0A1WWK2 A0A210QH45 A0A2M4ASF1 A0A0A1WN51 K1RB89 A0A182V1D5 A0A2J7QLT3 A0A1J1IBT4 A0A0L0C895 W5JIX0 A0A336M3H9 A0A182HQD2 A0A0R1DZM6 A0A1B6M7X2 A0A336LW49 A0A0A1WFV2 B4N5G0 A0A182P5V5 L7M8G8 B3NEF3 A0A131XYN6 A0A336LR78 A8E6R2 A0A1I8Q0P5 W8B5N7 V5GUJ9 A0A1S3IEV7 Q9VNR7 Q95TT3 A0A336LRV8 A0A0B7AD12 A0A293MBL7 A0A1A9VFA7 E9GEQ7 A0A224YT48 A0A1B0A2D4 B4PIP8 A0A131Z5C2 T1KR84 B3MB82 G3MK55 T1JJ22 Q29F13 H3A3S3 A0A1W4UR20 B4LDY2 A0A0C9RF91 A0A0J9RZY9
A0A0N1I7H1 A0A2H1VJE8 H9JPZ6 A0A226DS77 U5EHG1 A0A1Q3FIX3 Q17NN1 T1IEJ9 T1DLB3 A0A1Y9IUA8 A0A2R7VZ99 A0A023F4P8 A0A224XBX3 A0A0P4VHK7 A0A0L8FK67 A0A1D1VKF4 A0A1Y1KAE8 A0A1Q3FIG9 A0A2M4BL18 A0A2M4BLV8 A0A0K8THH4 A0A0K2TFI8 A0A1I8MSC5 A0A182WZT6 Q5TMY1 A0A182WEX6 A0A182JE15 A0A2C9GKX9 T1P817 T1PKC2 A0A1Q3FID0 A0A0K8U5Q3 A0A0K8WKD6 A0A1B6GM39 A0A1B6KHZ6 B0X3N0 B0WFQ7 A0A084WIX2 A0A1V9XS66 A0A182Y1G8 A0A182QW57 A0A2M4CYM1 Q17NN2 A0A182MYT1 A0A023GNL2 A0A1E1XFD1 A0A0A1WWK2 A0A210QH45 A0A2M4ASF1 A0A0A1WN51 K1RB89 A0A182V1D5 A0A2J7QLT3 A0A1J1IBT4 A0A0L0C895 W5JIX0 A0A336M3H9 A0A182HQD2 A0A0R1DZM6 A0A1B6M7X2 A0A336LW49 A0A0A1WFV2 B4N5G0 A0A182P5V5 L7M8G8 B3NEF3 A0A131XYN6 A0A336LR78 A8E6R2 A0A1I8Q0P5 W8B5N7 V5GUJ9 A0A1S3IEV7 Q9VNR7 Q95TT3 A0A336LRV8 A0A0B7AD12 A0A293MBL7 A0A1A9VFA7 E9GEQ7 A0A224YT48 A0A1B0A2D4 B4PIP8 A0A131Z5C2 T1KR84 B3MB82 G3MK55 T1JJ22 Q29F13 H3A3S3 A0A1W4UR20 B4LDY2 A0A0C9RF91 A0A0J9RZY9
PDB
3N5M
E-value=3.63634e-31,
Score=335
Ontologies
PATHWAY
00250
Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00260 Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00260 Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
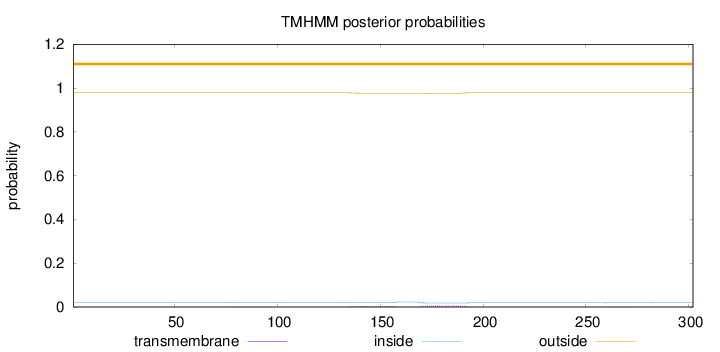
Length:
302
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01979
outside
1 - 302
Population Genetic Test Statistics
Pi
251.00807
Theta
185.100004
Tajima's D
1.190988
CLR
0
CSRT
0.713714314284286
Interpretation
Uncertain
