Gene
KWMTBOMO06425
Pre Gene Modal
BGIBMGA011949
Annotation
Sodium-dependent_neutral_amino_acid_transporter_B(0)AT2_[Papilio_machaon]
Full name
Transporter
Location in the cell
PlasmaMembrane Reliability : 4.952
Sequence
CDS
ATGGCGAACACAGCGCATTTGATGAGACGACAGAGTTCGCGGGACCTTTACGCGCAGAGGTCTCTGGATCGGATGGAGATGAAGGAGCTGAGAGGACGCCTCGTTACGATGGAGAACGGGCAGCCCGCGCACAGGACTCATGTCATGTACGGAGCTACTAATGAGGCCTTTGAAGACACAAGCCCGAACAGACGCTCCTCCGAAACTCCTACCAGCAAAGGGAGTTCCGGTGAAGAACGTGCCGGTTTTAAGAATGAGGGAGCCGAGATAGAACGCGAGTCTTGGGATAGTAAATTGACCTTCCTACTCGCAACCGTCGGATATGCTGTCGGCCTAGGAAACGTTTGGCGATTCCCATATTTAGCCCAAAAGAACGGCGGCGGTGCGTTTCTCATACCGTACTTCGTAATGCTGGCTATCGAGGGCATACCGATATTTTACTTGGAATTGGCAATCGGTCAGAGATTAAGGAAAGGCGCTATCGGCGTTTGGCATCAAGTATCCCCTTACCTCGGAGGTATCGGCATTAGTTCCGCCGTGGTTTCTTTCAACGTGGCTTTGTACTACAACTCTATAATAGCTTGGTGCCTTTTCTACTTCGCTCAGAGTTTCCAGGCGGAACTACCTTGGTCTGAATGCCCGAAGAAATACTTCCCGAACGGCTCTTACACAAGCGAACCGGAGTGCGCGAAAAGCAGCCCCACCCAATACTTTTGGTATCGAACAACTCTTCAAATATCCGAAGACATAAACTATCCGGAAACGTTCAATTACAAAATAGCTCTTGCTCTAGTCATTGCTTGGATTCTGGTCTACCTATGTATGATCAAAGGGATTGCGTCATCGGGGAAGGTAGTGTACGTGACCGCAACATTCCCATACATAGTCCTTGTTATATTTTTCTTTAGAGGCATCACATTGAAAGGCATGAGCGACGGACTAATACATCTATTCACTCCGAAGTGGCACACGATTTTAGACCCTGTGGTGTGGCTAGAAGCGGGCACACAGATCTTCTTTTCGCTTGGACTGGCTTTTGGTGGTTTAATAGCCTTCTCTTCGTACAACCCGGTCGATAACAACTGCTACAGAGACGCCATTATGGTCTCCATGACAAATTGTCTAACATCGATGTTCGCTGGTATCGTTGTCTTTTCTATAATTGGCTTCAAAGCAACTATGATCTATGAGAAATGTTTGGAGATCAGGAACGAGACTTTATTGGAGATGTTTGGCTCGTCATACAAGTCAATGGTGTTTCCTGAGGCTGGGGAATCGGTGACGATCGATAAAAACGGAACTATAACGAATTTGTTAATGCCACTATTGCCTGTTTGCGATTTGCAGAAGGAACTCGATAATACCGCATCCGGGACCGGTTTAGCGTTCATCATCTTCACCGAGGCCATTAACCAATTCCCAGGCGCTCAATTTTGGTCAGTCCTCTTCTTCCTGATGCTTTTCACCCTCGGGATCGATTCCCAGTTTGGGACCCTTGAGGGCGTAGTCACTTCCATCGTGGATATGAAGCTGTTCCCGAACCTGCGCAAGGAGTACCTGACCGGAGGGCTGTGCTTGCTCTGTTGCTTGCTGTCAATGGGCTTCGCTCATGGTGCAGGCAGCTATATATTCCTGCTCTTCGACATGTATAGCGGGAACTTCCCGCTACTGATCATTGCGTTCTTTGAATGCGTCGGGATCGCTTACGTGTACGGAGTGAAGAGGTGA
Protein
MANTAHLMRRQSSRDLYAQRSLDRMEMKELRGRLVTMENGQPAHRTHVMYGATNEAFEDTSPNRRSSETPTSKGSSGEERAGFKNEGAEIERESWDSKLTFLLATVGYAVGLGNVWRFPYLAQKNGGGAFLIPYFVMLAIEGIPIFYLELAIGQRLRKGAIGVWHQVSPYLGGIGISSAVVSFNVALYYNSIIAWCLFYFAQSFQAELPWSECPKKYFPNGSYTSEPECAKSSPTQYFWYRTTLQISEDINYPETFNYKIALALVIAWILVYLCMIKGIASSGKVVYVTATFPYIVLVIFFFRGITLKGMSDGLIHLFTPKWHTILDPVVWLEAGTQIFFSLGLAFGGLIAFSSYNPVDNNCYRDAIMVSMTNCLTSMFAGIVVFSIIGFKATMIYEKCLEIRNETLLEMFGSSYKSMVFPEAGESVTIDKNGTITNLLMPLLPVCDLQKELDNTASGTGLAFIIFTEAINQFPGAQFWSVLFFLMLFTLGIDSQFGTLEGVVTSIVDMKLFPNLRKEYLTGGLCLLCCLLSMGFAHGAGSYIFLLFDMYSGNFPLLIIAFFECVGIAYVYGVKR
Summary
Similarity
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Uniprot
A0A2H1VB97
A0A0N1PI46
A0A2A4J2I1
A0A2W1BYK9
A0A194PUT7
N6TH77
+ More
D6WF78 A0A3L8DFG3 F4WDU6 A0A067QK51 V9IIT8 A0A0M9A559 A0A087ZZD7 E2B634 A0A154PJZ4 E2AIM7 A0A232ES14 A0A2J7PQJ5 A0A195DUT1 A0A195FFX7 A0A195CRF3 A0A151XIK1 A0A026WYB0 E9IDL1 T1I2Z2 A0A195AW29 E0VLT1 A0A2R7WZI5 A0A1W4XVE7 A0A0J7L8S8 A0A2A3EK71 A0A0L7R0Y6 A0A084VQ96 A0A182RKR6 A0A1I8MX21 A0A182V1D1 A0A182TEQ0 A0A182IA05 A0A182M6Y4 A0A182Q4H7 A0A182W5V4 A0A182KAF6 Q29ID8 A0A3B0KLZ4 A0A1A9ULI3 A0A182FK95 B4M1X1 B4JIW6 A0A0L0C570 B7Z122 B4ND29 B3NSP8 A0A182GTQ1 B4Q0J8 A0A182YCR3 B4R458 A0A1I8P2J0 B3N065 W8BPP3 A0A182N0X1 A0A1A9W244 A0A1W4V8C1 B4L8L5 A0A0M4EXL4 A0A0P5AI26 A0A0P6JIK8 A0A182IQ63 A0A226EX01 A0A336MM67 A0A1A9X7Q0 A0A310SFP8 A0A182X7D4 A0A1B0AX17 E9G7X0 A0A1A9ZZD9 A0A0P6B6Q4 A0A1D2NGC4 A0A1B0FAE9 Q17AJ1 Q7PRN6 A0A0Q9WLC7 A0A182LDK5 Q9W4S0 A0A158NWR0 A0A0P5HCE5 A0A0P5C839 A0A1J1I9A8 K7J6L6 A0A1S4FAC8 Q8IRU1 A0A1V9XB45 B4I171 A0A0K2T2Z0 A0A0P5HQW6 T1ISA1 A0A210QQ71 K1P775 K1QJY2 A0A0L8HB76
D6WF78 A0A3L8DFG3 F4WDU6 A0A067QK51 V9IIT8 A0A0M9A559 A0A087ZZD7 E2B634 A0A154PJZ4 E2AIM7 A0A232ES14 A0A2J7PQJ5 A0A195DUT1 A0A195FFX7 A0A195CRF3 A0A151XIK1 A0A026WYB0 E9IDL1 T1I2Z2 A0A195AW29 E0VLT1 A0A2R7WZI5 A0A1W4XVE7 A0A0J7L8S8 A0A2A3EK71 A0A0L7R0Y6 A0A084VQ96 A0A182RKR6 A0A1I8MX21 A0A182V1D1 A0A182TEQ0 A0A182IA05 A0A182M6Y4 A0A182Q4H7 A0A182W5V4 A0A182KAF6 Q29ID8 A0A3B0KLZ4 A0A1A9ULI3 A0A182FK95 B4M1X1 B4JIW6 A0A0L0C570 B7Z122 B4ND29 B3NSP8 A0A182GTQ1 B4Q0J8 A0A182YCR3 B4R458 A0A1I8P2J0 B3N065 W8BPP3 A0A182N0X1 A0A1A9W244 A0A1W4V8C1 B4L8L5 A0A0M4EXL4 A0A0P5AI26 A0A0P6JIK8 A0A182IQ63 A0A226EX01 A0A336MM67 A0A1A9X7Q0 A0A310SFP8 A0A182X7D4 A0A1B0AX17 E9G7X0 A0A1A9ZZD9 A0A0P6B6Q4 A0A1D2NGC4 A0A1B0FAE9 Q17AJ1 Q7PRN6 A0A0Q9WLC7 A0A182LDK5 Q9W4S0 A0A158NWR0 A0A0P5HCE5 A0A0P5C839 A0A1J1I9A8 K7J6L6 A0A1S4FAC8 Q8IRU1 A0A1V9XB45 B4I171 A0A0K2T2Z0 A0A0P5HQW6 T1ISA1 A0A210QQ71 K1P775 K1QJY2 A0A0L8HB76
Pubmed
26354079
28756777
23537049
18362917
19820115
30249741
+ More
21719571 24845553 20798317 28648823 24508170 21282665 20566863 24438588 25315136 15632085 17994087 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26483478 17550304 25244985 24495485 21292972 27289101 17510324 12364791 20966253 26109357 26109356 21347285 20075255 28327890 28812685 22992520
21719571 24845553 20798317 28648823 24508170 21282665 20566863 24438588 25315136 15632085 17994087 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26483478 17550304 25244985 24495485 21292972 27289101 17510324 12364791 20966253 26109357 26109356 21347285 20075255 28327890 28812685 22992520
EMBL
ODYU01001388
SOQ37504.1
KQ460971
KPJ10142.1
NWSH01003679
PCG65946.1
+ More
KZ149896 PZC78735.1 KQ459590 KPI97171.1 APGK01027141 APGK01027142 KB740648 KB630489 KB631709 KB632148 ENN79789.1 ERL83626.1 ERL85521.1 ERL89103.1 KQ971319 EFA00300.1 QOIP01000008 RLU19205.1 GL888093 EGI67632.1 KK853359 KDR08170.1 JR049473 AEY60975.1 KQ435732 KOX77450.1 GL445914 EFN88823.1 KQ434938 KZC12143.1 GL439840 EFN66716.1 NNAY01002512 OXU21159.1 NEVH01022635 PNF18602.1 KQ980322 KYN16660.1 KQ981617 KYN39261.1 KQ977444 KYN02689.1 KQ982080 KYQ60118.1 KK107064 EZA60823.1 GL762454 EFZ21446.1 ACPB03006720 KQ976732 KYM76174.1 DS235281 EEB14337.1 KK856124 PTY24946.1 LBMM01000247 KMR03124.1 KZ288230 PBC31586.1 KQ414669 KOC64513.1 ATLV01015153 KE525003 KFB40140.1 APCN01001081 AXCM01002848 AXCN02001363 CH379063 EAL32715.3 OUUW01000011 SPP86856.1 CH940651 EDW65675.1 CH916370 EDV99530.1 JRES01000902 KNC27425.1 AE014298 ACL82883.1 CH964239 EDW82738.1 CH954180 EDV45728.1 JXUM01087655 KQ563649 KXJ73558.1 CM000162 EDX01282.1 CM000366 EDX17000.1 CH902640 EDV38269.1 GAMC01005628 JAC00928.1 CH933815 EDW07990.1 CP012528 ALC48517.1 GDIP01203826 LRGB01002076 JAJ19576.1 KZS09409.1 GDIQ01006874 JAN87863.1 LNIX01000001 OXA62069.1 UFQT01001722 SSX31522.1 KQ761738 OAD57131.1 JXJN01004969 GL732534 EFX84589.1 GDIP01018964 JAM84751.1 LJIJ01000048 ODN04311.1 CCAG010008637 CCAG010008638 CH477333 EAT43303.1 AAAB01008847 EAA06912.6 KRF82337.1 BT001443 AAN09096.1 AAN71198.1 ADTU01028380 GDIQ01229585 JAK22140.1 GDIP01174114 JAJ49288.1 CVRI01000044 CRK96853.1 AAZX01009864 AAF45873.3 MNPL01016780 OQR70656.1 CH480819 EDW53252.1 HACA01002586 CDW19947.1 GDIQ01226226 JAK25499.1 JH431429 NEDP02002420 OWF50883.1 JH818927 EKC19482.1 JH816138 EKC34058.1 KQ418612 KOF86531.1
KZ149896 PZC78735.1 KQ459590 KPI97171.1 APGK01027141 APGK01027142 KB740648 KB630489 KB631709 KB632148 ENN79789.1 ERL83626.1 ERL85521.1 ERL89103.1 KQ971319 EFA00300.1 QOIP01000008 RLU19205.1 GL888093 EGI67632.1 KK853359 KDR08170.1 JR049473 AEY60975.1 KQ435732 KOX77450.1 GL445914 EFN88823.1 KQ434938 KZC12143.1 GL439840 EFN66716.1 NNAY01002512 OXU21159.1 NEVH01022635 PNF18602.1 KQ980322 KYN16660.1 KQ981617 KYN39261.1 KQ977444 KYN02689.1 KQ982080 KYQ60118.1 KK107064 EZA60823.1 GL762454 EFZ21446.1 ACPB03006720 KQ976732 KYM76174.1 DS235281 EEB14337.1 KK856124 PTY24946.1 LBMM01000247 KMR03124.1 KZ288230 PBC31586.1 KQ414669 KOC64513.1 ATLV01015153 KE525003 KFB40140.1 APCN01001081 AXCM01002848 AXCN02001363 CH379063 EAL32715.3 OUUW01000011 SPP86856.1 CH940651 EDW65675.1 CH916370 EDV99530.1 JRES01000902 KNC27425.1 AE014298 ACL82883.1 CH964239 EDW82738.1 CH954180 EDV45728.1 JXUM01087655 KQ563649 KXJ73558.1 CM000162 EDX01282.1 CM000366 EDX17000.1 CH902640 EDV38269.1 GAMC01005628 JAC00928.1 CH933815 EDW07990.1 CP012528 ALC48517.1 GDIP01203826 LRGB01002076 JAJ19576.1 KZS09409.1 GDIQ01006874 JAN87863.1 LNIX01000001 OXA62069.1 UFQT01001722 SSX31522.1 KQ761738 OAD57131.1 JXJN01004969 GL732534 EFX84589.1 GDIP01018964 JAM84751.1 LJIJ01000048 ODN04311.1 CCAG010008637 CCAG010008638 CH477333 EAT43303.1 AAAB01008847 EAA06912.6 KRF82337.1 BT001443 AAN09096.1 AAN71198.1 ADTU01028380 GDIQ01229585 JAK22140.1 GDIP01174114 JAJ49288.1 CVRI01000044 CRK96853.1 AAZX01009864 AAF45873.3 MNPL01016780 OQR70656.1 CH480819 EDW53252.1 HACA01002586 CDW19947.1 GDIQ01226226 JAK25499.1 JH431429 NEDP02002420 OWF50883.1 JH818927 EKC19482.1 JH816138 EKC34058.1 KQ418612 KOF86531.1
Proteomes
UP000053240
UP000218220
UP000053268
UP000019118
UP000030742
UP000007266
+ More
UP000279307 UP000007755 UP000027135 UP000053105 UP000005203 UP000008237 UP000076502 UP000000311 UP000215335 UP000235965 UP000078492 UP000078541 UP000078542 UP000075809 UP000053097 UP000015103 UP000078540 UP000009046 UP000192223 UP000036403 UP000242457 UP000053825 UP000030765 UP000075900 UP000095301 UP000075903 UP000075902 UP000075840 UP000075883 UP000075886 UP000075920 UP000075881 UP000001819 UP000268350 UP000078200 UP000069272 UP000008792 UP000001070 UP000037069 UP000000803 UP000007798 UP000008711 UP000069940 UP000249989 UP000002282 UP000076408 UP000000304 UP000095300 UP000007801 UP000075884 UP000091820 UP000192221 UP000009192 UP000092553 UP000076858 UP000075880 UP000198287 UP000092443 UP000076407 UP000092460 UP000000305 UP000092445 UP000094527 UP000092444 UP000008820 UP000007062 UP000075882 UP000005205 UP000183832 UP000002358 UP000192247 UP000001292 UP000242188 UP000005408 UP000053454
UP000279307 UP000007755 UP000027135 UP000053105 UP000005203 UP000008237 UP000076502 UP000000311 UP000215335 UP000235965 UP000078492 UP000078541 UP000078542 UP000075809 UP000053097 UP000015103 UP000078540 UP000009046 UP000192223 UP000036403 UP000242457 UP000053825 UP000030765 UP000075900 UP000095301 UP000075903 UP000075902 UP000075840 UP000075883 UP000075886 UP000075920 UP000075881 UP000001819 UP000268350 UP000078200 UP000069272 UP000008792 UP000001070 UP000037069 UP000000803 UP000007798 UP000008711 UP000069940 UP000249989 UP000002282 UP000076408 UP000000304 UP000095300 UP000007801 UP000075884 UP000091820 UP000192221 UP000009192 UP000092553 UP000076858 UP000075880 UP000198287 UP000092443 UP000076407 UP000092460 UP000000305 UP000092445 UP000094527 UP000092444 UP000008820 UP000007062 UP000075882 UP000005205 UP000183832 UP000002358 UP000192247 UP000001292 UP000242188 UP000005408 UP000053454
Interpro
IPR037272
SNS_sf
+ More
IPR000175 Na/ntran_symport
IPR002438 Neutral_aa_SLC6
IPR029159 CK2S
IPR036322 WD40_repeat_dom_sf
IPR002181 Fibrinogen_a/b/g_C_dom
IPR001881 EGF-like_Ca-bd_dom
IPR036056 Fibrinogen-like_C
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR000742 EGF-like_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR000175 Na/ntran_symport
IPR002438 Neutral_aa_SLC6
IPR029159 CK2S
IPR036322 WD40_repeat_dom_sf
IPR002181 Fibrinogen_a/b/g_C_dom
IPR001881 EGF-like_Ca-bd_dom
IPR036056 Fibrinogen-like_C
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR000742 EGF-like_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
CDD
ProteinModelPortal
A0A2H1VB97
A0A0N1PI46
A0A2A4J2I1
A0A2W1BYK9
A0A194PUT7
N6TH77
+ More
D6WF78 A0A3L8DFG3 F4WDU6 A0A067QK51 V9IIT8 A0A0M9A559 A0A087ZZD7 E2B634 A0A154PJZ4 E2AIM7 A0A232ES14 A0A2J7PQJ5 A0A195DUT1 A0A195FFX7 A0A195CRF3 A0A151XIK1 A0A026WYB0 E9IDL1 T1I2Z2 A0A195AW29 E0VLT1 A0A2R7WZI5 A0A1W4XVE7 A0A0J7L8S8 A0A2A3EK71 A0A0L7R0Y6 A0A084VQ96 A0A182RKR6 A0A1I8MX21 A0A182V1D1 A0A182TEQ0 A0A182IA05 A0A182M6Y4 A0A182Q4H7 A0A182W5V4 A0A182KAF6 Q29ID8 A0A3B0KLZ4 A0A1A9ULI3 A0A182FK95 B4M1X1 B4JIW6 A0A0L0C570 B7Z122 B4ND29 B3NSP8 A0A182GTQ1 B4Q0J8 A0A182YCR3 B4R458 A0A1I8P2J0 B3N065 W8BPP3 A0A182N0X1 A0A1A9W244 A0A1W4V8C1 B4L8L5 A0A0M4EXL4 A0A0P5AI26 A0A0P6JIK8 A0A182IQ63 A0A226EX01 A0A336MM67 A0A1A9X7Q0 A0A310SFP8 A0A182X7D4 A0A1B0AX17 E9G7X0 A0A1A9ZZD9 A0A0P6B6Q4 A0A1D2NGC4 A0A1B0FAE9 Q17AJ1 Q7PRN6 A0A0Q9WLC7 A0A182LDK5 Q9W4S0 A0A158NWR0 A0A0P5HCE5 A0A0P5C839 A0A1J1I9A8 K7J6L6 A0A1S4FAC8 Q8IRU1 A0A1V9XB45 B4I171 A0A0K2T2Z0 A0A0P5HQW6 T1ISA1 A0A210QQ71 K1P775 K1QJY2 A0A0L8HB76
D6WF78 A0A3L8DFG3 F4WDU6 A0A067QK51 V9IIT8 A0A0M9A559 A0A087ZZD7 E2B634 A0A154PJZ4 E2AIM7 A0A232ES14 A0A2J7PQJ5 A0A195DUT1 A0A195FFX7 A0A195CRF3 A0A151XIK1 A0A026WYB0 E9IDL1 T1I2Z2 A0A195AW29 E0VLT1 A0A2R7WZI5 A0A1W4XVE7 A0A0J7L8S8 A0A2A3EK71 A0A0L7R0Y6 A0A084VQ96 A0A182RKR6 A0A1I8MX21 A0A182V1D1 A0A182TEQ0 A0A182IA05 A0A182M6Y4 A0A182Q4H7 A0A182W5V4 A0A182KAF6 Q29ID8 A0A3B0KLZ4 A0A1A9ULI3 A0A182FK95 B4M1X1 B4JIW6 A0A0L0C570 B7Z122 B4ND29 B3NSP8 A0A182GTQ1 B4Q0J8 A0A182YCR3 B4R458 A0A1I8P2J0 B3N065 W8BPP3 A0A182N0X1 A0A1A9W244 A0A1W4V8C1 B4L8L5 A0A0M4EXL4 A0A0P5AI26 A0A0P6JIK8 A0A182IQ63 A0A226EX01 A0A336MM67 A0A1A9X7Q0 A0A310SFP8 A0A182X7D4 A0A1B0AX17 E9G7X0 A0A1A9ZZD9 A0A0P6B6Q4 A0A1D2NGC4 A0A1B0FAE9 Q17AJ1 Q7PRN6 A0A0Q9WLC7 A0A182LDK5 Q9W4S0 A0A158NWR0 A0A0P5HCE5 A0A0P5C839 A0A1J1I9A8 K7J6L6 A0A1S4FAC8 Q8IRU1 A0A1V9XB45 B4I171 A0A0K2T2Z0 A0A0P5HQW6 T1ISA1 A0A210QQ71 K1P775 K1QJY2 A0A0L8HB76
PDB
6AWQ
E-value=7.05615e-93,
Score=871
Ontologies
GO
PANTHER
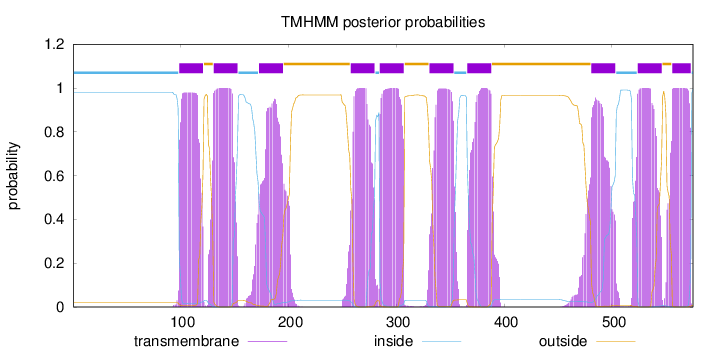
Topology
Length:
575
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
218.55792
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98049
inside
1 - 98
TMhelix
99 - 121
outside
122 - 130
TMhelix
131 - 153
inside
154 - 172
TMhelix
173 - 195
outside
196 - 257
TMhelix
258 - 280
inside
281 - 284
TMhelix
285 - 307
outside
308 - 330
TMhelix
331 - 353
inside
354 - 365
TMhelix
366 - 388
outside
389 - 480
TMhelix
481 - 503
inside
504 - 523
TMhelix
524 - 546
outside
547 - 555
TMhelix
556 - 573
inside
574 - 575
Population Genetic Test Statistics
Pi
233.626866
Theta
186.984902
Tajima's D
1.105975
CLR
0.569742
CSRT
0.684065796710165
Interpretation
Uncertain
