Gene
KWMTBOMO06419
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.543 Nuclear Reliability : 1.66
Sequence
CDS
ATGGACGCTGTATTCGCGGAATTCCTCCGACTTCGCCACCCACAGCTCGCCTCGGAGTTTTTGGCCTTCAAGGCCAATCACACTGCGAGCCCTCTCGAGGACTCCGCCGCGCTCGCTACTCCTACGTCGCCTGTACCTGCGTGCAGAGCTTCTGCATTGAGCACCGCAGCCTCTGTCGTGCCTGCCGCTCCCGTGTCGCCTATACTGGCGAGAAAAGCTGCTGCGTCGTCCGCCGTGACCATCGTTTCAGCTGAGCGATCATCCGCGGCCTCCGTCGCGCCCTCTAAAACACCTACACTTGCTCGTAGGTCGCCTGCACCCGCCTCCTCGTGCTCCGACTCTGACTCGGACATGGAGGTCGACCTCGCCCCCGCCTCATCGACGGATGGATTCACCCTGGTACAGAAGGGTAAGAAGCGTGCCGCGGAGTCTCGAGCTCCCGCGGCCGCTAAAATTAGCAAAGCCGTGAACGCGTCGCGCCCCCGCCCTCAGACTCCCGTTGCGCCCCCAGCCCGTGCCACTCCGTCGCCGCGTCCGGTGGCACAAAATAAAACCCAGACCCCTCCCCCGGTTATCCTTCAGGAGAAGGCAGCTTGGGATCGAGTTTGCCTGGCCCTTAAGGCCAAAAATATAAATTTCACGAATGCCCGTAACCTCGCGAACGGCATTCAAATTAAGGTTCAAACACCCGACGACCATAGGGCCCTCTCTTCTTACCTCCGTAAGGAGCGTATAAGTTTCCATACGTATACGCTCCAGGAGGAGCGCGAACTCCGCGTTGTAATACGCGGAATCCCTAAAGAGTTAGATGTAGAGCTCGTAAAAGCCGACCTGTTAGAACAAGGCCTACCAGTGAATTCTGTGCACCGTATGCACACCGGTCGCGGTAGGGAGCCATATAATATGGTTCTAGTCGCTCTCCAGCCTACCCCCGAGGTGCAGGAGACCCTACTTAAGCCCGCGCGCCGTGACCCTAAAATCGCGAACTATAACATGGTCAGGAACGACAGGCTCTCTGCCCGTGGTGGTGGTACCGTCATTTACTATAGAAGAGCCCTGCATTGCGTCCCGCTCGATCCTCCCGCGCTCGCTAATATCGAAGCATCAGTGTGCCGAATCTCACTGACGGGACACGCGCCGATCGTTATCGCGTCCGTTTATCTTCCACCGGATAAGATCGTTCTAAGCAGTGATATCGAGGCGCTGCTCGGTATGGGGAGCTCTGTCATTCTGGCGGGCGACCTAAATTGTAAACACATCAGGTGGAACTCACACACCACAACCCCGAATGGCAGGCGGCTTGACGCGTTAGTCGATGATCTCGCCTTCGATATCGTCGCTCCGCTAACCCCGACTCACTACCCGCTAAATATCGCGCATCGCCCGGATATACTCGACATAGCGTTATTAAAAAACGTAACTCTGCGCTTACACTCGATCGAAGTAGTTTCAGAGTTAGATTCAGACCACCGTCCCGTCGTTATGAAGCTCGGTCGCGCTCCCGATTCCGTTCCCGTCACGAGGACTGTGGTGGATTGGCACACGCTGGGCATCAGCCTGGCTGAATCTGATCCACCATCGCTCCCGTTTAACCCGGACTCTATCCCGTCTCCTCAGGATACCGCTGAAGCCATAGACATCTTAACGTCACACATCACCTCGACATTAGATAGGTCATCGAAACAAGTTGTAGAGGAGGACTTCCTTCACCGCTTCAAATTGTCCGACGATATTAGGGAACTTCTTAGAGCTAAGAACGCCTCGATACGCGCCTACGACAGGTATCCTACCGCGGAAAATCGTATTCGAATGCGTGCCCTACAACGCGACGTAAAGTCTCGCATCGCCGAAGTCCGAGATGCCAGATGGTCTGATTTCTTAGAAGGACTCGCGCCCTCCCAAAGGTCTTACTACCGCTTAGCTCGTACTCTCAAATCGGATACGGTAGTAACTATGCCCCCCCTCGTAGGCCCCTCAGGCCGACTCGCGGCGTTCGATGATGACGAAAAAGCAGAGCTGCTGGCCGATACATTGCAAACCCAGTGCACGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGCCTCCTTGCCACCCTCTGATGCGTTACCACCCGTCACCCCGATGGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACGGTATATCCAACCGCGTTATTAAACTTCTACCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGCATACATAAACCCGGTAAACCAAAAAATCATCCGACGAGCTACCGCCCGATTAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGCGTCTGCTCTACAAACGCCTCAGAGACTTCGTCTCATCCAAGGGCATTCTTATCGATGAACAATTCGGATTCCGTACAAATCACTCATGCGTTCAACAGGTGCACCGCCTCACGGAGCACATTCTTGTAGGGCTTAATCGACCAAAACCGTTATACACGGGAGCTCTCTTCTTCGACGTCGCAAAAGCGTTCGACAAAGTCTGGCACAATGGTTTGATTTTCAAACTATTCAACATGGGCGTGCCGGATAGTCTCGTGCTCATCATACGGGACTTCTTGTCGAACCGCTCTTTTCGATATCGAGTCGAGGGAACCCGCTCCTCCCCACGACCTCTCACAGCTGGAGTCCCGCAAGGCTCTGTCCTCTCACCCCTCCTATTTAGCTTATTCGTCAACGATATTCCCCGGTCGCCGCCGACCCATTTAGCTTTATTCGCCGACGACACGACTGTTTACTATTCTAGTAGAAATAAGTCCCTAATCGCGAAGAAGCTTCAGAGCGCAGCCCTAGCCCTAGGACAGTGGTTCCGAAAATGGCGCATAGACATCAACCCAGCGAAAAGTACTGCGGTGCTATTTCAGAGGGAAGCTCCACACGGATTTCCTCCCGGATTAGGAGGAGGAATCTCACACCCCCGATTACTCTCTTTAGACAACCCATACCCTGGGCCAGGAAGCTGTCGGGCTCCGTGGTTCGTGAGGAACGTTGACCTACACGACGACCTGGGCCTCGAATCAATTCGGAAATACATGAAGTCAGCGTCGGAACGATACTTCGATAAGGCTATGCGTCATGATAATCGCCTTATAGTTGCCGCCGCTGACTACTCCCCGAATCCTGATCATGCAGGAGCCAGTCACCGTCGACGCCCTAGACACGTCCTTACGGATCCATCAGATCCAATAACCTTTGCATTAGATGCCTTCAGCTCTAATACTAGGGGCAGGCTTAGGGACCCCGATTTGCAGATAGATCCTATGTCCCGTGTAGTGTTCCACCATGAAGGAATACAATTAATTCTTCGACCTCAAACAACTGCAATCACGTCCTAG
Protein
MDAVFAEFLRLRHPQLASEFLAFKANHTASPLEDSAALATPTSPVPACRASALSTAASVVPAAPVSPILARKAAASSAVTIVSAERSSAASVAPSKTPTLARRSPAPASSCSDSDSDMEVDLAPASSTDGFTLVQKGKKRAAESRAPAAAKISKAVNASRPRPQTPVAPPARATPSPRPVAQNKTQTPPPVILQEKAAWDRVCLALKAKNINFTNARNLANGIQIKVQTPDDHRALSSYLRKERISFHTYTLQEERELRVVIRGIPKELDVELVKADLLEQGLPVNSVHRMHTGRGREPYNMVLVALQPTPEVQETLLKPARRDPKIANYNMVRNDRLSARGGGTVIYYRRALHCVPLDPPALANIEASVCRISLTGHAPIVIASVYLPPDKIVLSSDIEALLGMGSSVILAGDLNCKHIRWNSHTTTPNGRRLDALVDDLAFDIVAPLTPTHYPLNIAHRPDILDIALLKNVTLRLHSIEVVSELDSDHRPVVMKLGRAPDSVPVTRTVVDWHTLGISLAESDPPSLPFNPDSIPSPQDTAEAIDILTSHITSTLDRSSKQVVEEDFLHRFKLSDDIRELLRAKNASIRAYDRYPTAENRIRMRALQRDVKSRIAEVRDARWSDFLEGLAPSQRSYYRLARTLKSDTVVTMPPLVGPSGRLAAFDDDEKAELLADTLQTQCTPSTQSVDPVHVELVDSEVERRASLPPSDALPPVTPMEVKDLIKDLRPRKAPGSDGISNRVIKLLPVQLIVMLASIFNAAMANCIFPAVWKEADVIGIHKPGKPKNHPTSYRPISLLMSLGKLYERLLYKRLRDFVSSKGILIDEQFGFRTNHSCVQQVHRLTEHILVGLNRPKPLYTGALFFDVAKAFDKVWHNGLIFKLFNMGVPDSLVLIIRDFLSNRSFRYRVEGTRSSPRPLTAGVPQGSVLSPLLFSLFVNDIPRSPPTHLALFADDTTVYYSSRNKSLIAKKLQSAALALGQWFRKWRIDINPAKSTAVLFQREAPHGFPPGLGGGISHPRLLSLDNPYPGPGSCRAPWFVRNVDLHDDLGLESIRKYMKSASERYFDKAMRHDNRLIVAAADYSPNPDHAGASHRRRPRHVLTDPSDPITFALDAFSSNTRGRLRDPDLQIDPMSRVVFHHEGIQLILRPQTTAITS
Summary
Uniprot
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
Ontologies
GO
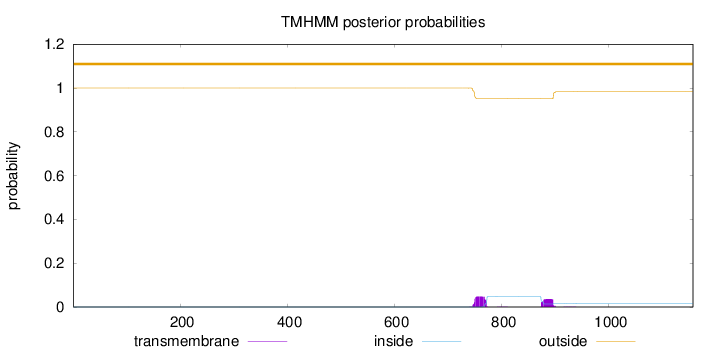
Topology
Length:
1157
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.87811
Exp number, first 60 AAs:
0.00104
Total prob of N-in:
0.00014
outside
1 - 1157
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
119.297034
CSRT
0
Interpretation
Uncertain
