Gene
KWMTBOMO06414
Pre Gene Modal
BGIBMGA011953
Annotation
PREDICTED:_uncharacterized_protein_LOC101738118_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.052
Sequence
CDS
ATGAGGGTTGAAGAAAAATCATTCGTTCGTGAAAATTTTCAAGCAAACATAACCAACTGTAACGAATTTCCTCCGTCACTTGAAAGACACATTCGAAATATTATAATTAAGGAAGGCTATACTAAATATAGTGTAGATAAAAGAGAAATATGCACAAATGGTGGAAACTATCTCGCTTCGCTTTACGAAATCGACATAAAAGGTACAACACAAAAAGGAAAAAAAGAAACGAACATTTTCATTAAGAAAATGAAGCCGGGCGAAAATAGTATGAAAATTATATCAATACCAGAAGCATATAAATTCGAAACATTTGCCTATGAAGAATTGTTAAAGGTATATACAGAACTGCAAAATGAAGCTAAAGTACCTGCCGATGAAAGATTCAAAGCGGTAAAAGATTTTGGGGAATTTGATGCTCAAGCTATCATACTCGAAAATTTAGCGAAAAAAGGATATAAAACTGCTCACAGGATGGACGTTGTGTCTTTGGAATACGCTGAAATGGCAATCAAGCAATTAGCAAAATTCCATGCTTTATCTTTTGCTTTACGGGAAAGAAAGCCGGAATATTATGAATCTAAAATTAAGACGATCCCATCGCGTTTGAATCTAGGAAACGAATGGAATGGTTTCGTAAAGTCAATGCAAGCTTTAACTTTGAATTTTTTGAATGATAACTTGAGGAAGAAAGTGGAGAATATTTTGCCGCGTTTATTCGATAATATAACGGATTATTTTGAAGGAAATGATTCAGCTGTATGTTGTTTGTGTCACGGAGATTTTAGACCTAATAACATTATGATGAAAGAAATAGATGGTGAAATATCAGAAACAATTCCAATCGATTACCAACTGATATACTATGGATGTCCAATAATAGATTTCTTATACTTCGTATTCACGGCAACCGACAAAGAATTCAGAAGAGCTCATTTAACACACCTGAAAACCCTATATTACGATACAATGCAAAATTTTCTTGAATATTTTGATATTTCTATAGAATCCGTGTATTCTAAAACAGATTTCGATAATGATTATAAGAAAAAACTTGATTACGGTTTAATTGTTACCATTTTGTATTGCCCCTTTATGTTTGTGGACGAGAACGACGTACCCGACATTACTGAAGCTGATCTAGGCAATATATCGTTTACTTTACATGATAAATACAAAGATAAAATCCAGGGAACTGTTGAGGACTTCATTGAATGGGGATTAATTTAA
Protein
MRVEEKSFVRENFQANITNCNEFPPSLERHIRNIIIKEGYTKYSVDKREICTNGGNYLASLYEIDIKGTTQKGKKETNIFIKKMKPGENSMKIISIPEAYKFETFAYEELLKVYTELQNEAKVPADERFKAVKDFGEFDAQAIILENLAKKGYKTAHRMDVVSLEYAEMAIKQLAKFHALSFALRERKPEYYESKIKTIPSRLNLGNEWNGFVKSMQALTLNFLNDNLRKKVENILPRLFDNITDYFEGNDSAVCCLCHGDFRPNNIMMKEIDGEISETIPIDYQLIYYGCPIIDFLYFVFTATDKEFRRAHLTHLKTLYYDTMQNFLEYFDISIESVYSKTDFDNDYKKKLDYGLIVTILYCPFMFVDENDVPDITEADLGNISFTLHDKYKDKIQGTVEDFIEWGLI
Summary
Uniprot
H9JQZ6
A0A2W1BZN5
A0A2A4K7V6
A0A2H1VB73
A0A0L7LNU4
A0A194PVZ6
+ More
A0A2W1C180 A0A0N0PDU1 A0A0N0PDI8 A0A194PVY1 A0A212ESF8 A0A194PV19 H9JQZ7 A0A3S2LZ05 A0A3S2NXY2 A0A0N1IB22 A0A3S2TJ07 A0A212FDY3 A0A2A4K7M3 A0A3S2N2X9 A0A1E1WHV5 A0A212EQU8 A0A0N1I9M5 A0A0N1PGW5 A0A2A4J644 Q0PCR8 A0A0N1PEQ1 A0A2A4JJ09 A0A2A4JJH8 A0A212F2J2 A0A2A4JFI8 A0A2A4JU06 A0A0L7KXT2 A0A2A4JV59 A0A2A4JF80 A0A2A4JE13 A0A0N0PB04 A0A0S1MMB6 H9JSW8 A0A2W1BTX3 A0A2A4JDZ5 A0A2W1BG42 A0A1Z2RRI0 A0A212EL62 A0A2W1BSP2 A0A212FGC4 A0A0N1IMS0 A0A2A4JTG5 A0A2W1BRE8 A0A0L7KX70 A0A0N0PAP6 A0A0L7LPX6 A0A2A4JJJ2 A0A212FGF5 A0A2W1BRI8 A0A212EH39 A0A2A4JGC4 A0A3S2TJN1 A0A2W1BUA5 A0A2H1V9I6 A0A2H1V9N0 A0A0N1I2U1 A0A212EPE1 A0A0L7KXI3 A0A2H1V9H4 A0A0N1PFY2 A0A2H1VB90 A0A2W1BMZ2 A0A212EPI2 A0A212FKY4 A0A2H1WA95 A0A0L7LQZ0 A0A0A1XLB8 A0A1B0G905 A0A2W1BJJ0 Q7PXT1 A0A182L9K2 A0A336KVV2 A0A2A4JDV8 A0A336L2B2 A0A1A9UQR7 B0XJW2 A0A2C9GQM2 B4MKV4 A0A0L0BQ72 A0A1A9Z2Y6 A0A182VHT4 A0A2C9GQM4 A0A0N0PAP1 A0A1A9WRH8 A0A1A9Y035 A0A1B0B3L1 A0A1Q3G4V1 A0A182WSB5 F5HMM0
A0A2W1C180 A0A0N0PDU1 A0A0N0PDI8 A0A194PVY1 A0A212ESF8 A0A194PV19 H9JQZ7 A0A3S2LZ05 A0A3S2NXY2 A0A0N1IB22 A0A3S2TJ07 A0A212FDY3 A0A2A4K7M3 A0A3S2N2X9 A0A1E1WHV5 A0A212EQU8 A0A0N1I9M5 A0A0N1PGW5 A0A2A4J644 Q0PCR8 A0A0N1PEQ1 A0A2A4JJ09 A0A2A4JJH8 A0A212F2J2 A0A2A4JFI8 A0A2A4JU06 A0A0L7KXT2 A0A2A4JV59 A0A2A4JF80 A0A2A4JE13 A0A0N0PB04 A0A0S1MMB6 H9JSW8 A0A2W1BTX3 A0A2A4JDZ5 A0A2W1BG42 A0A1Z2RRI0 A0A212EL62 A0A2W1BSP2 A0A212FGC4 A0A0N1IMS0 A0A2A4JTG5 A0A2W1BRE8 A0A0L7KX70 A0A0N0PAP6 A0A0L7LPX6 A0A2A4JJJ2 A0A212FGF5 A0A2W1BRI8 A0A212EH39 A0A2A4JGC4 A0A3S2TJN1 A0A2W1BUA5 A0A2H1V9I6 A0A2H1V9N0 A0A0N1I2U1 A0A212EPE1 A0A0L7KXI3 A0A2H1V9H4 A0A0N1PFY2 A0A2H1VB90 A0A2W1BMZ2 A0A212EPI2 A0A212FKY4 A0A2H1WA95 A0A0L7LQZ0 A0A0A1XLB8 A0A1B0G905 A0A2W1BJJ0 Q7PXT1 A0A182L9K2 A0A336KVV2 A0A2A4JDV8 A0A336L2B2 A0A1A9UQR7 B0XJW2 A0A2C9GQM2 B4MKV4 A0A0L0BQ72 A0A1A9Z2Y6 A0A182VHT4 A0A2C9GQM4 A0A0N0PAP1 A0A1A9WRH8 A0A1A9Y035 A0A1B0B3L1 A0A1Q3G4V1 A0A182WSB5 F5HMM0
Pubmed
EMBL
BABH01028544
KZ149896
PZC78727.1
NWSH01000065
PCG80018.1
ODYU01001618
+ More
SOQ38093.1 JTDY01000433 KOB77193.1 KQ459590 KPI97168.1 PZC78726.1 KQ460210 KPJ16578.1 KPJ16577.1 KPI97153.1 AGBW02012788 OWR44436.1 KPI97167.1 RSAL01000104 RVE47388.1 RVE47386.1 KPJ16591.1 RVE47387.1 AGBW02008998 OWR51897.1 PCG80016.1 RSAL01004670 RVE40111.1 GDQN01004485 JAT86569.1 AGBW02013226 OWR43849.1 KQ459449 KPJ00881.1 KQ461184 KPJ07528.1 NWSH01003122 PCG66910.1 AB252485 BAF02792.1 KPJ00882.1 NWSH01001354 PCG71564.1 PCG71563.1 AGBW02010741 OWR47943.1 NWSH01001667 PCG70468.1 NWSH01000613 PCG75299.1 JTDY01004714 KOB67844.1 PCG75300.1 PCG70469.1 NWSH01001934 PCG69633.1 KPJ07527.1 KR821066 ALL42054.1 BABH01036018 BABH01036019 KZ149965 PZC76230.1 PCG69632.1 KZ150068 PZC74089.1 KY938812 ASA46453.1 AGBW02014137 OWR42208.1 PZC76227.1 AGBW02008687 OWR52782.1 KPJ00871.1 PCG75301.1 PZC76224.1 KOB67847.1 KPJ07536.1 JTDY01000356 KOB77568.1 PCG71562.1 OWR52783.1 PZC76225.1 AGBW02014964 OWR40805.1 PCG70470.1 RSAL01000092 RVE47952.1 PZC76226.1 ODYU01001386 SOQ37495.1 SOQ37496.1 KPJ00870.1 AGBW02013474 OWR43365.1 KOB67845.1 SOQ37500.1 KPJ00880.1 SOQ37494.1 PZC74090.1 OWR43367.1 AGBW02007938 OWR54408.1 ODYU01007302 SOQ49967.1 JTDY01000292 KOB77867.1 GBXI01017142 GBXI01002969 JAC97149.1 JAD11323.1 CCAG010002956 KZ150144 PZC72910.1 AAAB01008987 EAA00815.5 UFQS01001072 UFQT01001072 SSX08839.1 SSX28751.1 NWSH01001948 PCG69592.1 UFQS01001320 UFQT01001320 SSX10331.1 SSX30019.1 DS233624 EDS30841.1 APCN01000881 CH963847 EDW72879.2 JRES01001542 KNC22156.1 KPJ07529.1 JXJN01007892 GFDL01000217 JAV34828.1 EGK97542.1
SOQ38093.1 JTDY01000433 KOB77193.1 KQ459590 KPI97168.1 PZC78726.1 KQ460210 KPJ16578.1 KPJ16577.1 KPI97153.1 AGBW02012788 OWR44436.1 KPI97167.1 RSAL01000104 RVE47388.1 RVE47386.1 KPJ16591.1 RVE47387.1 AGBW02008998 OWR51897.1 PCG80016.1 RSAL01004670 RVE40111.1 GDQN01004485 JAT86569.1 AGBW02013226 OWR43849.1 KQ459449 KPJ00881.1 KQ461184 KPJ07528.1 NWSH01003122 PCG66910.1 AB252485 BAF02792.1 KPJ00882.1 NWSH01001354 PCG71564.1 PCG71563.1 AGBW02010741 OWR47943.1 NWSH01001667 PCG70468.1 NWSH01000613 PCG75299.1 JTDY01004714 KOB67844.1 PCG75300.1 PCG70469.1 NWSH01001934 PCG69633.1 KPJ07527.1 KR821066 ALL42054.1 BABH01036018 BABH01036019 KZ149965 PZC76230.1 PCG69632.1 KZ150068 PZC74089.1 KY938812 ASA46453.1 AGBW02014137 OWR42208.1 PZC76227.1 AGBW02008687 OWR52782.1 KPJ00871.1 PCG75301.1 PZC76224.1 KOB67847.1 KPJ07536.1 JTDY01000356 KOB77568.1 PCG71562.1 OWR52783.1 PZC76225.1 AGBW02014964 OWR40805.1 PCG70470.1 RSAL01000092 RVE47952.1 PZC76226.1 ODYU01001386 SOQ37495.1 SOQ37496.1 KPJ00870.1 AGBW02013474 OWR43365.1 KOB67845.1 SOQ37500.1 KPJ00880.1 SOQ37494.1 PZC74090.1 OWR43367.1 AGBW02007938 OWR54408.1 ODYU01007302 SOQ49967.1 JTDY01000292 KOB77867.1 GBXI01017142 GBXI01002969 JAC97149.1 JAD11323.1 CCAG010002956 KZ150144 PZC72910.1 AAAB01008987 EAA00815.5 UFQS01001072 UFQT01001072 SSX08839.1 SSX28751.1 NWSH01001948 PCG69592.1 UFQS01001320 UFQT01001320 SSX10331.1 SSX30019.1 DS233624 EDS30841.1 APCN01000881 CH963847 EDW72879.2 JRES01001542 KNC22156.1 KPJ07529.1 JXJN01007892 GFDL01000217 JAV34828.1 EGK97542.1
Proteomes
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JQZ6
A0A2W1BZN5
A0A2A4K7V6
A0A2H1VB73
A0A0L7LNU4
A0A194PVZ6
+ More
A0A2W1C180 A0A0N0PDU1 A0A0N0PDI8 A0A194PVY1 A0A212ESF8 A0A194PV19 H9JQZ7 A0A3S2LZ05 A0A3S2NXY2 A0A0N1IB22 A0A3S2TJ07 A0A212FDY3 A0A2A4K7M3 A0A3S2N2X9 A0A1E1WHV5 A0A212EQU8 A0A0N1I9M5 A0A0N1PGW5 A0A2A4J644 Q0PCR8 A0A0N1PEQ1 A0A2A4JJ09 A0A2A4JJH8 A0A212F2J2 A0A2A4JFI8 A0A2A4JU06 A0A0L7KXT2 A0A2A4JV59 A0A2A4JF80 A0A2A4JE13 A0A0N0PB04 A0A0S1MMB6 H9JSW8 A0A2W1BTX3 A0A2A4JDZ5 A0A2W1BG42 A0A1Z2RRI0 A0A212EL62 A0A2W1BSP2 A0A212FGC4 A0A0N1IMS0 A0A2A4JTG5 A0A2W1BRE8 A0A0L7KX70 A0A0N0PAP6 A0A0L7LPX6 A0A2A4JJJ2 A0A212FGF5 A0A2W1BRI8 A0A212EH39 A0A2A4JGC4 A0A3S2TJN1 A0A2W1BUA5 A0A2H1V9I6 A0A2H1V9N0 A0A0N1I2U1 A0A212EPE1 A0A0L7KXI3 A0A2H1V9H4 A0A0N1PFY2 A0A2H1VB90 A0A2W1BMZ2 A0A212EPI2 A0A212FKY4 A0A2H1WA95 A0A0L7LQZ0 A0A0A1XLB8 A0A1B0G905 A0A2W1BJJ0 Q7PXT1 A0A182L9K2 A0A336KVV2 A0A2A4JDV8 A0A336L2B2 A0A1A9UQR7 B0XJW2 A0A2C9GQM2 B4MKV4 A0A0L0BQ72 A0A1A9Z2Y6 A0A182VHT4 A0A2C9GQM4 A0A0N0PAP1 A0A1A9WRH8 A0A1A9Y035 A0A1B0B3L1 A0A1Q3G4V1 A0A182WSB5 F5HMM0
A0A2W1C180 A0A0N0PDU1 A0A0N0PDI8 A0A194PVY1 A0A212ESF8 A0A194PV19 H9JQZ7 A0A3S2LZ05 A0A3S2NXY2 A0A0N1IB22 A0A3S2TJ07 A0A212FDY3 A0A2A4K7M3 A0A3S2N2X9 A0A1E1WHV5 A0A212EQU8 A0A0N1I9M5 A0A0N1PGW5 A0A2A4J644 Q0PCR8 A0A0N1PEQ1 A0A2A4JJ09 A0A2A4JJH8 A0A212F2J2 A0A2A4JFI8 A0A2A4JU06 A0A0L7KXT2 A0A2A4JV59 A0A2A4JF80 A0A2A4JE13 A0A0N0PB04 A0A0S1MMB6 H9JSW8 A0A2W1BTX3 A0A2A4JDZ5 A0A2W1BG42 A0A1Z2RRI0 A0A212EL62 A0A2W1BSP2 A0A212FGC4 A0A0N1IMS0 A0A2A4JTG5 A0A2W1BRE8 A0A0L7KX70 A0A0N0PAP6 A0A0L7LPX6 A0A2A4JJJ2 A0A212FGF5 A0A2W1BRI8 A0A212EH39 A0A2A4JGC4 A0A3S2TJN1 A0A2W1BUA5 A0A2H1V9I6 A0A2H1V9N0 A0A0N1I2U1 A0A212EPE1 A0A0L7KXI3 A0A2H1V9H4 A0A0N1PFY2 A0A2H1VB90 A0A2W1BMZ2 A0A212EPI2 A0A212FKY4 A0A2H1WA95 A0A0L7LQZ0 A0A0A1XLB8 A0A1B0G905 A0A2W1BJJ0 Q7PXT1 A0A182L9K2 A0A336KVV2 A0A2A4JDV8 A0A336L2B2 A0A1A9UQR7 B0XJW2 A0A2C9GQM2 B4MKV4 A0A0L0BQ72 A0A1A9Z2Y6 A0A182VHT4 A0A2C9GQM4 A0A0N0PAP1 A0A1A9WRH8 A0A1A9Y035 A0A1B0B3L1 A0A1Q3G4V1 A0A182WSB5 F5HMM0
Ontologies
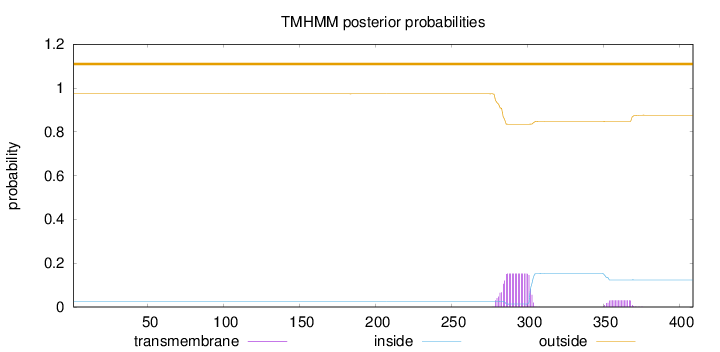
Topology
Length:
409
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.57772999999999
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.02643
outside
1 - 409
Population Genetic Test Statistics
Pi
229.925198
Theta
204.782734
Tajima's D
0.386494
CLR
0
CSRT
0.47997600119994
Interpretation
Uncertain
