Pre Gene Modal
BGIBMGA011939
Annotation
putative_bisphosphate_nucleotidase_[Danaus_plexippus]
Full name
Putative inositol monophosphatase 3
Alternative Name
Inositol-1(or 4)-monophosphatase 3
Myo-inositol monophosphatase A3
Myo-inositol monophosphatase A3
Location in the cell
Cytoplasmic Reliability : 1.268 Extracellular Reliability : 1.527
Sequence
CDS
ATGAATCTCGGTGGGACATTACGAATTAACAAGTTCGCTTGTTTCACATTGGCTTTTGTATTATTTCTGATAATATACTGGCGATCCGGAACATCTAGCTATCCTACAGAGAAGAGTCACTTAGTTAATTTAAAGAGTTTATTAAAAGCCGCTATCTTAGCCGCCGAAGGAGGAGGCAAGAAAGTATTGGACGGAAAAAATCATATTTTGAATGTTAAGAGTAAGGGTAAAACTTTGGAAGGCGCCAACGACCCTGTAACTGATGTTGATTATGCATCGCATTGCGCTATGTATTATGGTTTAAAACACACTTTTAATAATATCGATGTGGTCTCTGAGGAGCATTCGAAGAGTGATAGTAGCTGCGAGGAGGTGCCTGCTTTCGACTTGGAGCCCGACATCCCCGGGAATCTGAACATTGAGCAAATGCTTGATGAGCTTGTGCTTGCTAAAGATGTTGTTGTTTGGATCGACCCATTAGATGCCACCCAGGAGTATACAGAGAAACTGTACCAATATGTTACAACAATGGTATGTGTTGCTATAAGAGGAGTGCCAATAATAGGAGTCATCCATTATCCATTCACACAGCAAACATATTGGGCATGGTATACTAAGAAAACATCAGCTAATATGCCACTTGTACCGCATAAAGAAGAAAACAGGGAACATCCTAGAGTGGTAATTTCTCGATCACATCCTGGAAAAGTTGCCACTGTAGCCAAAAGTGCTTTTGGAGACAAGTCCAATGTTTCCAAAGCTGCTGGAGCAGGATTCAAGCAGATGGCCATGTTGCAGTGA
Protein
MNLGGTLRINKFACFTLAFVLFLIIYWRSGTSSYPTEKSHLVNLKSLLKAAILAAEGGGKKVLDGKNHILNVKSKGKTLEGANDPVTDVDYASHCAMYYGLKHTFNNIDVVSEEHSKSDSSCEEVPAFDLEPDIPGNLNIEQMLDELVLAKDVVVWIDPLDATQEYTEKLYQYVTTMVCVAIRGVPIIGVIHYPFTQQTYWAWYTKKTSANMPLVPHKEENREHPRVVISRSHPGKVATVAKSAFGDKSNVSKAAGAGFKQMAMLQ
Summary
Catalytic Activity
a myo-inositol phosphate + H2O = myo-inositol + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the inositol monophosphatase superfamily.
Keywords
Complete proteome
Hydrolase
Magnesium
Membrane
Metal-binding
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Putative inositol monophosphatase 3
Uniprot
H9JQY3
A0A2H1VVT3
A0A2W1BUF5
A0A2A4J047
A0A3S2NSG7
A0A212FDZ6
+ More
A0A0L7KPB0 A0A0N1PHB7 A0A194PUS7 Q17CJ1 A0A1S4F800 A0A023ERE4 B0W8L8 A0A1L8DM98 A0A1Q3FCW4 A0A1B0GGR1 A0A3Q0J0B0 A0A1B0DDA0 A0A182SQD0 A0A182YGF7 A0A182RNH2 A0A0A9WGF4 A0A182VR88 A0A182VGG8 A0A182UD35 A0A182HKR5 A0A182X9T3 A0A182L928 A0A1J1I3D2 A0A2M4AVM2 A0A2M4AW41 A0A0V0G7I0 Q7PXG6 A0A182Q4E6 A0A182N5D2 A0A182JPB6 A0A023F0T6 A0A182P2H2 A0A182F794 A0A2M4BSV3 W5JIG5 A0A2M4BT85 A0A336KC29 E0VYW2 A0A2J7PCX7 A0A224XS35 W8BXB6 A0A182IUH4 A0A084WQT5 A0A2B4RX26 A0A1Y1KLA4 A0A0P4WBY4 A0A0P4WKP8 A0A3M6TB15 A0A182GPE3 T1I5R4 U5ESQ0 A0A1I8N884 T1P9S2 A0A2G8KJE7 A0A1I8NMX7 A0A1B6L012 A0A182MC89 A0A0A1X8R0 A0A069DT15 A0A034VSD0 A0A0K8VC75 A0A293MA12 A0A293LFI8 A0A2R7W4F9 D3TM44 Q29JH0 A0A1A9VS92 A0A2P8XM43 A0A1A9YA74 A0A3F2Z4P6 T2M4I0 A0A1B6GM15 A0A1Z5LAN6 B3NWQ7 B4Q233 A0A1W4VK88 B4M7C2 M9PH13 Q9VYF2 A0A1A9WVA1 A0A154P0E1 A0A087UY50 B4IGQ1 A0A067R5J1 D6WH25 A0A3B0J6V8 W4XS15 A0A310SM18 B4JJ71 A0A2P6K4G3 A0A2T7NNW6 A0A0L7QXB1 A0A0M9AE81 B7PTD3
A0A0L7KPB0 A0A0N1PHB7 A0A194PUS7 Q17CJ1 A0A1S4F800 A0A023ERE4 B0W8L8 A0A1L8DM98 A0A1Q3FCW4 A0A1B0GGR1 A0A3Q0J0B0 A0A1B0DDA0 A0A182SQD0 A0A182YGF7 A0A182RNH2 A0A0A9WGF4 A0A182VR88 A0A182VGG8 A0A182UD35 A0A182HKR5 A0A182X9T3 A0A182L928 A0A1J1I3D2 A0A2M4AVM2 A0A2M4AW41 A0A0V0G7I0 Q7PXG6 A0A182Q4E6 A0A182N5D2 A0A182JPB6 A0A023F0T6 A0A182P2H2 A0A182F794 A0A2M4BSV3 W5JIG5 A0A2M4BT85 A0A336KC29 E0VYW2 A0A2J7PCX7 A0A224XS35 W8BXB6 A0A182IUH4 A0A084WQT5 A0A2B4RX26 A0A1Y1KLA4 A0A0P4WBY4 A0A0P4WKP8 A0A3M6TB15 A0A182GPE3 T1I5R4 U5ESQ0 A0A1I8N884 T1P9S2 A0A2G8KJE7 A0A1I8NMX7 A0A1B6L012 A0A182MC89 A0A0A1X8R0 A0A069DT15 A0A034VSD0 A0A0K8VC75 A0A293MA12 A0A293LFI8 A0A2R7W4F9 D3TM44 Q29JH0 A0A1A9VS92 A0A2P8XM43 A0A1A9YA74 A0A3F2Z4P6 T2M4I0 A0A1B6GM15 A0A1Z5LAN6 B3NWQ7 B4Q233 A0A1W4VK88 B4M7C2 M9PH13 Q9VYF2 A0A1A9WVA1 A0A154P0E1 A0A087UY50 B4IGQ1 A0A067R5J1 D6WH25 A0A3B0J6V8 W4XS15 A0A310SM18 B4JJ71 A0A2P6K4G3 A0A2T7NNW6 A0A0L7QXB1 A0A0M9AE81 B7PTD3
EC Number
3.1.3.25
Pubmed
19121390
28756777
22118469
26227816
26354079
17510324
+ More
24945155 25244985 25401762 26823975 20966253 12364791 14747013 17210077 25474469 20920257 23761445 20566863 24495485 24438588 28004739 30382153 26483478 25315136 29023486 25830018 26334808 25348373 20353571 15632085 29403074 24065732 28528879 17994087 17550304 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 24845553 18362917 19820115
24945155 25244985 25401762 26823975 20966253 12364791 14747013 17210077 25474469 20920257 23761445 20566863 24495485 24438588 28004739 30382153 26483478 25315136 29023486 25830018 26334808 25348373 20353571 15632085 29403074 24065732 28528879 17994087 17550304 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 24845553 18362917 19820115
EMBL
BABH01028565
ODYU01004580
SOQ44602.1
KZ149897
PZC78678.1
NWSH01004140
+ More
PCG65445.1 RSAL01000104 RVE47377.1 AGBW02008998 OWR51907.1 JTDY01007781 KOB64945.1 KQ460210 KPJ16584.1 KQ459590 KPI97161.1 CH477308 EAT44030.1 GAPW01002359 JAC11239.1 DS231859 EDS39068.1 GFDF01006600 JAV07484.1 GFDL01009717 JAV25328.1 AJWK01000607 AJVK01031805 GBHO01040945 GBHO01037123 GBRD01000688 GDHC01019060 GDHC01007824 JAG02659.1 JAG06481.1 JAG65133.1 JAP99568.1 JAQ10805.1 APCN01000860 CVRI01000038 CRK94100.1 GGFK01011518 MBW44839.1 GGFK01011601 MBW44922.1 GECL01002118 JAP04006.1 AAAB01008987 EAA01450.4 AXCN02001179 GBBI01003622 JAC15090.1 GGFJ01006999 MBW56140.1 ADMH02001070 ETN64182.1 GGFJ01007000 MBW56141.1 UFQS01000188 UFQT01000188 SSX00985.1 SSX21365.1 DS235848 EEB18568.1 NEVH01026401 PNF14189.1 GFTR01005176 JAW11250.1 GAMC01000605 JAC05951.1 ATLV01025724 KE525398 KFB52579.1 LSMT01000299 PFX20878.1 GEZM01084874 GEZM01084872 GEZM01084871 GEZM01084870 JAV60445.1 GDRN01082559 JAI61811.1 GDRN01082560 JAI61810.1 RCHS01003977 RMX38587.1 JXUM01078327 JXUM01078328 JXUM01078329 JXUM01078330 JXUM01078331 JXUM01078332 JXUM01078333 KQ563059 KXJ74557.1 ACPB03006895 GANO01002256 JAB57615.1 KA645501 AFP60130.1 MRZV01000539 PIK48123.1 GEBQ01022935 JAT17042.1 AXCM01008432 GBXI01006603 JAD07689.1 GBGD01002082 JAC86807.1 GAKP01014258 GAKP01014257 JAC44695.1 GDHF01015836 JAI36478.1 GFWV01011241 MAA35970.1 GFWV01001908 MAA26638.1 KK854314 PTY14552.1 CCAG010020716 EZ422496 ADD18772.1 CH379063 PYGN01001739 PSN33073.1 JXJN01022690 HAAD01000734 CDG66966.1 GECZ01006283 JAS63486.1 GFJQ02002508 JAW04462.1 CH954180 EDV47219.1 CM000162 EDX01554.1 CH940653 EDW62689.2 KRF80762.1 AE014298 AGB95353.1 AHN59667.1 BT004861 KQ434783 KZC04824.1 KK122233 KFM82289.1 CH480836 EDW49001.1 KK852729 KDR17547.1 KQ971321 EFA00621.1 OUUW01000003 SPP77777.1 AAGJ04023973 KQ759893 OAD62071.1 CH916370 EDV99623.1 MWRG01030100 PRD21219.1 PZQS01000010 PVD22870.1 KQ414705 KOC63245.1 KQ435687 KOX81349.1 ABJB010610209 ABJB010735492 ABJB010778331 DS785108 EEC09855.1
PCG65445.1 RSAL01000104 RVE47377.1 AGBW02008998 OWR51907.1 JTDY01007781 KOB64945.1 KQ460210 KPJ16584.1 KQ459590 KPI97161.1 CH477308 EAT44030.1 GAPW01002359 JAC11239.1 DS231859 EDS39068.1 GFDF01006600 JAV07484.1 GFDL01009717 JAV25328.1 AJWK01000607 AJVK01031805 GBHO01040945 GBHO01037123 GBRD01000688 GDHC01019060 GDHC01007824 JAG02659.1 JAG06481.1 JAG65133.1 JAP99568.1 JAQ10805.1 APCN01000860 CVRI01000038 CRK94100.1 GGFK01011518 MBW44839.1 GGFK01011601 MBW44922.1 GECL01002118 JAP04006.1 AAAB01008987 EAA01450.4 AXCN02001179 GBBI01003622 JAC15090.1 GGFJ01006999 MBW56140.1 ADMH02001070 ETN64182.1 GGFJ01007000 MBW56141.1 UFQS01000188 UFQT01000188 SSX00985.1 SSX21365.1 DS235848 EEB18568.1 NEVH01026401 PNF14189.1 GFTR01005176 JAW11250.1 GAMC01000605 JAC05951.1 ATLV01025724 KE525398 KFB52579.1 LSMT01000299 PFX20878.1 GEZM01084874 GEZM01084872 GEZM01084871 GEZM01084870 JAV60445.1 GDRN01082559 JAI61811.1 GDRN01082560 JAI61810.1 RCHS01003977 RMX38587.1 JXUM01078327 JXUM01078328 JXUM01078329 JXUM01078330 JXUM01078331 JXUM01078332 JXUM01078333 KQ563059 KXJ74557.1 ACPB03006895 GANO01002256 JAB57615.1 KA645501 AFP60130.1 MRZV01000539 PIK48123.1 GEBQ01022935 JAT17042.1 AXCM01008432 GBXI01006603 JAD07689.1 GBGD01002082 JAC86807.1 GAKP01014258 GAKP01014257 JAC44695.1 GDHF01015836 JAI36478.1 GFWV01011241 MAA35970.1 GFWV01001908 MAA26638.1 KK854314 PTY14552.1 CCAG010020716 EZ422496 ADD18772.1 CH379063 PYGN01001739 PSN33073.1 JXJN01022690 HAAD01000734 CDG66966.1 GECZ01006283 JAS63486.1 GFJQ02002508 JAW04462.1 CH954180 EDV47219.1 CM000162 EDX01554.1 CH940653 EDW62689.2 KRF80762.1 AE014298 AGB95353.1 AHN59667.1 BT004861 KQ434783 KZC04824.1 KK122233 KFM82289.1 CH480836 EDW49001.1 KK852729 KDR17547.1 KQ971321 EFA00621.1 OUUW01000003 SPP77777.1 AAGJ04023973 KQ759893 OAD62071.1 CH916370 EDV99623.1 MWRG01030100 PRD21219.1 PZQS01000010 PVD22870.1 KQ414705 KOC63245.1 KQ435687 KOX81349.1 ABJB010610209 ABJB010735492 ABJB010778331 DS785108 EEC09855.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000037510
UP000053240
+ More
UP000053268 UP000008820 UP000002320 UP000092461 UP000079169 UP000092462 UP000075901 UP000076408 UP000075900 UP000075920 UP000075903 UP000075902 UP000075840 UP000076407 UP000075882 UP000183832 UP000007062 UP000075886 UP000075884 UP000075881 UP000075885 UP000069272 UP000000673 UP000009046 UP000235965 UP000075880 UP000030765 UP000225706 UP000275408 UP000069940 UP000249989 UP000015103 UP000095301 UP000230750 UP000095300 UP000075883 UP000092444 UP000001819 UP000078200 UP000245037 UP000092443 UP000092460 UP000008711 UP000002282 UP000192221 UP000008792 UP000000803 UP000091820 UP000076502 UP000054359 UP000001292 UP000027135 UP000007266 UP000268350 UP000007110 UP000001070 UP000245119 UP000053825 UP000053105 UP000001555
UP000053268 UP000008820 UP000002320 UP000092461 UP000079169 UP000092462 UP000075901 UP000076408 UP000075900 UP000075920 UP000075903 UP000075902 UP000075840 UP000076407 UP000075882 UP000183832 UP000007062 UP000075886 UP000075884 UP000075881 UP000075885 UP000069272 UP000000673 UP000009046 UP000235965 UP000075880 UP000030765 UP000225706 UP000275408 UP000069940 UP000249989 UP000015103 UP000095301 UP000230750 UP000095300 UP000075883 UP000092444 UP000001819 UP000078200 UP000245037 UP000092443 UP000092460 UP000008711 UP000002282 UP000192221 UP000008792 UP000000803 UP000091820 UP000076502 UP000054359 UP000001292 UP000027135 UP000007266 UP000268350 UP000007110 UP000001070 UP000245119 UP000053825 UP000053105 UP000001555
Interpro
SUPFAM
SSF54495
SSF54495
Gene 3D
CDD
ProteinModelPortal
H9JQY3
A0A2H1VVT3
A0A2W1BUF5
A0A2A4J047
A0A3S2NSG7
A0A212FDZ6
+ More
A0A0L7KPB0 A0A0N1PHB7 A0A194PUS7 Q17CJ1 A0A1S4F800 A0A023ERE4 B0W8L8 A0A1L8DM98 A0A1Q3FCW4 A0A1B0GGR1 A0A3Q0J0B0 A0A1B0DDA0 A0A182SQD0 A0A182YGF7 A0A182RNH2 A0A0A9WGF4 A0A182VR88 A0A182VGG8 A0A182UD35 A0A182HKR5 A0A182X9T3 A0A182L928 A0A1J1I3D2 A0A2M4AVM2 A0A2M4AW41 A0A0V0G7I0 Q7PXG6 A0A182Q4E6 A0A182N5D2 A0A182JPB6 A0A023F0T6 A0A182P2H2 A0A182F794 A0A2M4BSV3 W5JIG5 A0A2M4BT85 A0A336KC29 E0VYW2 A0A2J7PCX7 A0A224XS35 W8BXB6 A0A182IUH4 A0A084WQT5 A0A2B4RX26 A0A1Y1KLA4 A0A0P4WBY4 A0A0P4WKP8 A0A3M6TB15 A0A182GPE3 T1I5R4 U5ESQ0 A0A1I8N884 T1P9S2 A0A2G8KJE7 A0A1I8NMX7 A0A1B6L012 A0A182MC89 A0A0A1X8R0 A0A069DT15 A0A034VSD0 A0A0K8VC75 A0A293MA12 A0A293LFI8 A0A2R7W4F9 D3TM44 Q29JH0 A0A1A9VS92 A0A2P8XM43 A0A1A9YA74 A0A3F2Z4P6 T2M4I0 A0A1B6GM15 A0A1Z5LAN6 B3NWQ7 B4Q233 A0A1W4VK88 B4M7C2 M9PH13 Q9VYF2 A0A1A9WVA1 A0A154P0E1 A0A087UY50 B4IGQ1 A0A067R5J1 D6WH25 A0A3B0J6V8 W4XS15 A0A310SM18 B4JJ71 A0A2P6K4G3 A0A2T7NNW6 A0A0L7QXB1 A0A0M9AE81 B7PTD3
A0A0L7KPB0 A0A0N1PHB7 A0A194PUS7 Q17CJ1 A0A1S4F800 A0A023ERE4 B0W8L8 A0A1L8DM98 A0A1Q3FCW4 A0A1B0GGR1 A0A3Q0J0B0 A0A1B0DDA0 A0A182SQD0 A0A182YGF7 A0A182RNH2 A0A0A9WGF4 A0A182VR88 A0A182VGG8 A0A182UD35 A0A182HKR5 A0A182X9T3 A0A182L928 A0A1J1I3D2 A0A2M4AVM2 A0A2M4AW41 A0A0V0G7I0 Q7PXG6 A0A182Q4E6 A0A182N5D2 A0A182JPB6 A0A023F0T6 A0A182P2H2 A0A182F794 A0A2M4BSV3 W5JIG5 A0A2M4BT85 A0A336KC29 E0VYW2 A0A2J7PCX7 A0A224XS35 W8BXB6 A0A182IUH4 A0A084WQT5 A0A2B4RX26 A0A1Y1KLA4 A0A0P4WBY4 A0A0P4WKP8 A0A3M6TB15 A0A182GPE3 T1I5R4 U5ESQ0 A0A1I8N884 T1P9S2 A0A2G8KJE7 A0A1I8NMX7 A0A1B6L012 A0A182MC89 A0A0A1X8R0 A0A069DT15 A0A034VSD0 A0A0K8VC75 A0A293MA12 A0A293LFI8 A0A2R7W4F9 D3TM44 Q29JH0 A0A1A9VS92 A0A2P8XM43 A0A1A9YA74 A0A3F2Z4P6 T2M4I0 A0A1B6GM15 A0A1Z5LAN6 B3NWQ7 B4Q233 A0A1W4VK88 B4M7C2 M9PH13 Q9VYF2 A0A1A9WVA1 A0A154P0E1 A0A087UY50 B4IGQ1 A0A067R5J1 D6WH25 A0A3B0J6V8 W4XS15 A0A310SM18 B4JJ71 A0A2P6K4G3 A0A2T7NNW6 A0A0L7QXB1 A0A0M9AE81 B7PTD3
PDB
1JP4
E-value=1.40897e-08,
Score=140
Ontologies
PATHWAY
GO
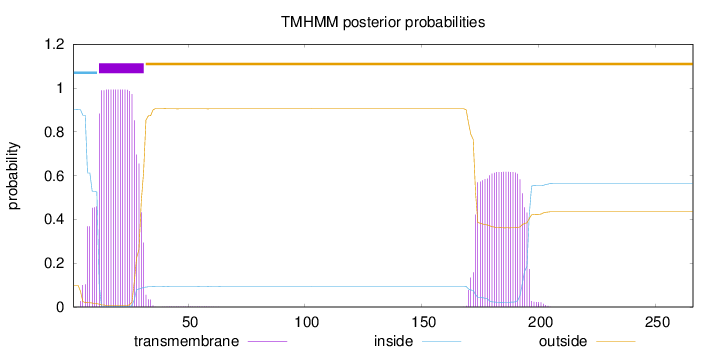
Topology
Subcellular location
Membrane
Length:
266
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
34.16291
Exp number, first 60 AAs:
20.18639
Total prob of N-in:
0.90129
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 31
outside
32 - 266
Population Genetic Test Statistics
Pi
137.306922
Theta
149.599124
Tajima's D
-0.473165
CLR
0.318318
CSRT
0.249137543122844
Interpretation
Uncertain
