Gene
KWMTBOMO06403
Pre Gene Modal
BGIBMGA011937
Annotation
PREDICTED:_tektin-B1-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.429
Sequence
CDS
ATGGACACATCAGTTCCAGTTTTTGAGAAAACTCATCCTCGTTTAACAGTTTTAGATTGGACTAGGAATATTCAAAAGTTACAAAATGAGGCAAGACACCGGAGGTTCGAGTCGTACGAATTACGACAAAGAGCTAATCAATTGCGCAATGAAACATGTATTACTACGAGATGGGATACTTACGTTAATGATGAACTATTAAGAGACAGAATCTTCGAGGTACAATCATGGCGTGAAAAGCAAAGATTCACACGGGATTGCGTGCGAGATGAGATCAGAGCTTTAAAGGAGGAGAAGCATGCAACAGAGTTACACTTAGAGTCACTCCAGGTGCCGCTCATGATAGTTTCACAGTGCCTGTCTAACAGGGACCAGAGAGTTCCCCCTGAGCTCACTAGAGATCAGCTAGAAGAAGAATTGAAGAAGGAGCTTCACATAATTGAGAATAGCAAACGAGTCCTGAAAGATGTATGCAACATGGGCTGGGAGAAGATTAAAGATCTAACCAACGTGCTCTGTAGACTGGAGAGGGAGATCCGAAACAAGGACGAGACGCTAGATTTGGAGTATCACGTCAAAGACTTGACTAGAGACAGCTCCGATATTTCATTTAAGCTTGATCCTACCAGGATACCTCCTCATACTATAACAGAAATAAATTACGTCGAATGCATAGAAAATACAGTGAAGATAGCAGAAAGTCTCATGTCCGAGTCAAAGAACCTTCGGGAGACGATGTTTAGGAGCCGAGAACAGGCCCAAAACCAGATGTACGCGCAGTGCCAGACTGTTGAAATGGTGATGAGGAGACGGGTTTATGATATACAGAGGGGGAGGAATGAGATGGATTGGCAGAAATACAAGCTAGAAGCGAACATGCAGAAAGTTGATCGAGAGATCGAGTCTCTTCGAGCAGCGGTCGCTGACAAAATAAACCCAACTAAACTGGTCGAAACTAGGCTGGAGACCAGGACTAGAAGACCTGTGCTTGAAAGAGTTGATGATAAACCTATGCGAGGGCTAATAGAAGAATTCGAAAGAGTGCACCTGAGTCATAGCAAACTTGAGAAGAAACTCGAAGACGCATTAACAACGTACCACGGCATGTACAATCACCATGAACGCCTCACCCGGGACCTGCAGCACAAGAACCAGGCTCTGGAGACGGACCGACGGCTCATAGAGATCCGGAAACCCTTGCATGCACCCGACGACACACAGTTCAAGAGGAACGTTGGCTATTGTCACATGAAGGACGAACTAGTTCCCGAGTAA
Protein
MDTSVPVFEKTHPRLTVLDWTRNIQKLQNEARHRRFESYELRQRANQLRNETCITTRWDTYVNDELLRDRIFEVQSWREKQRFTRDCVRDEIRALKEEKHATELHLESLQVPLMIVSQCLSNRDQRVPPELTRDQLEEELKKELHIIENSKRVLKDVCNMGWEKIKDLTNVLCRLEREIRNKDETLDLEYHVKDLTRDSSDISFKLDPTRIPPHTITEINYVECIENTVKIAESLMSESKNLRETMFRSREQAQNQMYAQCQTVEMVMRRRVYDIQRGRNEMDWQKYKLEANMQKVDREIESLRAAVADKINPTKLVETRLETRTRRPVLERVDDKPMRGLIEEFERVHLSHSKLEKKLEDALTTYHGMYNHHERLTRDLQHKNQALETDRRLIEIRKPLHAPDDTQFKRNVGYCHMKDELVPE
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. Protein arginine N-methyltransferase family.
Uniprot
H9JQY1
A0A2H1W3Z6
A0A2A4J1A4
A0A3S2NCB3
A0A0N1IPW0
A0A194PWM4
+ More
A0A212FDQ3 A0A2W1C0N9 A0A0L7LLC7 A0A2H1W8M1 A0A0N1II92 A0A336LSN8 A0A194QAF5 A0A182QK68 A0A212F0U9 A0A1W4WGQ3 A0A182RME7 A0A182JQ50 A0A084VY33 A0A182Y3G4 Q7Q482 A0A182MPW5 A0A182SLW7 A0A182MYZ6 A0A182XL58 A0A182V0R4 A0A182L0X1 A0A182ICG6 A0A1Q3FTE8 B0X6Z4 W5J667 A0A182J7Q2 A0A182PCP8 A0A182F4B3 A0A182H7Y6 A0A1Y1N046 A0A182W6P8 A0A182GIL1 A0A2A4J8U6 Q0IFA2 B0X6Z5 A0A1B6GWJ6 A0A182TRF2 A0A1B6CBT4 A0A1I8Q999 Q291Y4 B4GA29 A0A3B0JE42 A0A034W5F2 B4MNL2 W8BMQ2 A0A2P8YAE4 A0A0K8WAA7 A0A1B0D9Q4 B3MF30 B4KSL7 B4LPB5 D1GYC6 A0A1A9XJY6 B4JWL6 A0A1W4VRL2 A0A1B0AJC5 N6TKH4 A0A0L0CD00 B4P924 A0A2J7R9K0 D1GYC9 Q9W1V2 Q8I1F3 A0AMS9 U4TYH0 A0A1B0GCZ6 A0A0A9XWS8 B4LPB4 D6WGQ5 A0A1I8NF91 A0A0M4EH02 A0A0L7QY25 A0A2J7R9H9 D1GZ29 A0A0M9A8X8 A0A0J9RJE3 A0AMS6 B4QI54 B4I8G1 R4FQC8 T1IED1 A0A1B0CQM3 A0A1J1HE31 A0A1B6KR36 H9J9U6 A0A154PAW6 A0A224XCZ4 B4JWL7 A0A2A3E430 A0A087ZQP0 A0A2R7WU91 A0A195CB88 F4WY84 B4LPB3
A0A212FDQ3 A0A2W1C0N9 A0A0L7LLC7 A0A2H1W8M1 A0A0N1II92 A0A336LSN8 A0A194QAF5 A0A182QK68 A0A212F0U9 A0A1W4WGQ3 A0A182RME7 A0A182JQ50 A0A084VY33 A0A182Y3G4 Q7Q482 A0A182MPW5 A0A182SLW7 A0A182MYZ6 A0A182XL58 A0A182V0R4 A0A182L0X1 A0A182ICG6 A0A1Q3FTE8 B0X6Z4 W5J667 A0A182J7Q2 A0A182PCP8 A0A182F4B3 A0A182H7Y6 A0A1Y1N046 A0A182W6P8 A0A182GIL1 A0A2A4J8U6 Q0IFA2 B0X6Z5 A0A1B6GWJ6 A0A182TRF2 A0A1B6CBT4 A0A1I8Q999 Q291Y4 B4GA29 A0A3B0JE42 A0A034W5F2 B4MNL2 W8BMQ2 A0A2P8YAE4 A0A0K8WAA7 A0A1B0D9Q4 B3MF30 B4KSL7 B4LPB5 D1GYC6 A0A1A9XJY6 B4JWL6 A0A1W4VRL2 A0A1B0AJC5 N6TKH4 A0A0L0CD00 B4P924 A0A2J7R9K0 D1GYC9 Q9W1V2 Q8I1F3 A0AMS9 U4TYH0 A0A1B0GCZ6 A0A0A9XWS8 B4LPB4 D6WGQ5 A0A1I8NF91 A0A0M4EH02 A0A0L7QY25 A0A2J7R9H9 D1GZ29 A0A0M9A8X8 A0A0J9RJE3 A0AMS6 B4QI54 B4I8G1 R4FQC8 T1IED1 A0A1B0CQM3 A0A1J1HE31 A0A1B6KR36 H9J9U6 A0A154PAW6 A0A224XCZ4 B4JWL7 A0A2A3E430 A0A087ZQP0 A0A2R7WU91 A0A195CB88 F4WY84 B4LPB3
Pubmed
19121390
26354079
22118469
28756777
26227816
24438588
+ More
25244985 12364791 14747013 17210077 20966253 20920257 23761445 26483478 28004739 17510324 15632085 17994087 25348373 24495485 29403074 23537049 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 16951084 17569856 17569867 19126864 26109357 26109356 18057021 25401762 18362917 19820115 25315136 22936249 21719571
25244985 12364791 14747013 17210077 20966253 20920257 23761445 26483478 28004739 17510324 15632085 17994087 25348373 24495485 29403074 23537049 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 16951084 17569856 17569867 19126864 26109357 26109356 18057021 25401762 18362917 19820115 25315136 22936249 21719571
EMBL
BABH01028569
ODYU01006180
SOQ47811.1
NWSH01004140
PCG65448.1
RSAL01000104
+ More
RVE47374.1 KQ460210 KPJ16585.1 KQ459590 KPI97159.1 AGBW02008998 OWR51905.1 KZ149897 PZC78680.1 JTDY01000680 KOB76247.1 ODYU01007032 SOQ49418.1 KQ460324 KPJ15851.1 UFQT01000102 SSX20011.1 KQ459249 KPJ02414.1 AXCN02000789 AGBW02011024 OWR47360.1 ATLV01018264 KE525226 KFB42877.1 AAAB01008964 EAA12295.1 AXCM01002698 APCN01000798 GFDL01004227 JAV30818.1 DS232430 EDS41665.1 ADMH02002138 ETN58355.1 JXUM01117605 KQ566100 KXJ70398.1 GEZM01016394 JAV91291.1 JXUM01066478 KQ562400 KXJ75996.1 NWSH01002602 PCG67880.1 CH477389 EAT41984.1 EDS41666.1 GECZ01002937 JAS66832.1 GEDC01026380 JAS10918.1 CM000071 EAL24978.2 CH479181 EDW31781.1 OUUW01000001 SPP73530.1 GAKP01009425 JAC49527.1 CH963848 EDW73701.1 GAMC01011899 JAB94656.1 PYGN01000757 PSN41225.1 GDHF01004223 JAI48091.1 AJVK01028397 CH902619 EDV37658.1 CH933808 EDW10516.1 CH940648 EDW60224.1 FN546515 FN546516 FN546517 FN546519 FN546520 FN546521 FN546522 FN546523 FN546524 FN546525 CBE66814.1 CBE66815.1 CBE66816.1 CBE66818.1 CBE66819.1 CBE66820.1 CBE66821.1 CBE66822.1 CBE66823.1 CBE66824.1 CH916375 EDV98354.1 APGK01021190 KB740293 ENN80949.1 JRES01000577 KNC30125.1 CM000158 EDW92264.1 NEVH01006584 PNF37503.1 FN546518 CBE66817.1 AE013599 AY058393 AM293965 AM293966 AM293968 AM293969 AM293970 AM293972 AM293973 AM293974 AM293975 FM245093 FM245094 FM245095 FM245096 FM245097 FM245098 FM245099 FM245100 FM245101 FM245102 FM245103 FM245104 AAF46951.1 AAL13622.1 CAL25867.1 CAL25868.1 CAL25870.1 CAL25871.1 CAL25872.1 CAL25874.1 CAL25875.1 CAL25876.1 CAL25877.1 CAR93019.1 CAR93020.1 CAR93021.1 CAR93022.1 CAR93023.1 CAR93024.1 CAR93025.1 CAR93026.1 CAR93027.1 CAR93028.1 CAR93029.1 CAR93030.1 AY190941 CH954179 AAO01015.1 EDV56742.1 AM293967 AM293971 CAL25869.1 CAL25873.1 KB630005 KB630385 ERL83336.1 ERL83578.1 CCAG010002248 GBHO01021894 GBRD01003021 JAG21710.1 JAG62800.1 EDW60223.1 KQ971327 EEZ99614.1 CP012524 ALC41772.1 KQ414699 KOC63456.1 PNF37500.1 FN546768 CBE67067.1 KQ435724 KOX78353.1 CM002911 KMY95956.1 AM293964 CAL25866.1 CM000362 EDX08308.1 CH480824 EDW56886.1 GAHY01000469 JAA77041.1 ACPB03014062 AJWK01023760 CVRI01000001 CRK86224.1 GEBQ01026283 JAT13694.1 BABH01025880 KQ434846 KZC08368.1 GFTR01006131 JAW10295.1 EDV98355.1 KZ288386 PBC26487.1 KK855555 PTY23106.1 KQ978023 KYM98067.1 GL888439 EGI60841.1 EDW60222.2
RVE47374.1 KQ460210 KPJ16585.1 KQ459590 KPI97159.1 AGBW02008998 OWR51905.1 KZ149897 PZC78680.1 JTDY01000680 KOB76247.1 ODYU01007032 SOQ49418.1 KQ460324 KPJ15851.1 UFQT01000102 SSX20011.1 KQ459249 KPJ02414.1 AXCN02000789 AGBW02011024 OWR47360.1 ATLV01018264 KE525226 KFB42877.1 AAAB01008964 EAA12295.1 AXCM01002698 APCN01000798 GFDL01004227 JAV30818.1 DS232430 EDS41665.1 ADMH02002138 ETN58355.1 JXUM01117605 KQ566100 KXJ70398.1 GEZM01016394 JAV91291.1 JXUM01066478 KQ562400 KXJ75996.1 NWSH01002602 PCG67880.1 CH477389 EAT41984.1 EDS41666.1 GECZ01002937 JAS66832.1 GEDC01026380 JAS10918.1 CM000071 EAL24978.2 CH479181 EDW31781.1 OUUW01000001 SPP73530.1 GAKP01009425 JAC49527.1 CH963848 EDW73701.1 GAMC01011899 JAB94656.1 PYGN01000757 PSN41225.1 GDHF01004223 JAI48091.1 AJVK01028397 CH902619 EDV37658.1 CH933808 EDW10516.1 CH940648 EDW60224.1 FN546515 FN546516 FN546517 FN546519 FN546520 FN546521 FN546522 FN546523 FN546524 FN546525 CBE66814.1 CBE66815.1 CBE66816.1 CBE66818.1 CBE66819.1 CBE66820.1 CBE66821.1 CBE66822.1 CBE66823.1 CBE66824.1 CH916375 EDV98354.1 APGK01021190 KB740293 ENN80949.1 JRES01000577 KNC30125.1 CM000158 EDW92264.1 NEVH01006584 PNF37503.1 FN546518 CBE66817.1 AE013599 AY058393 AM293965 AM293966 AM293968 AM293969 AM293970 AM293972 AM293973 AM293974 AM293975 FM245093 FM245094 FM245095 FM245096 FM245097 FM245098 FM245099 FM245100 FM245101 FM245102 FM245103 FM245104 AAF46951.1 AAL13622.1 CAL25867.1 CAL25868.1 CAL25870.1 CAL25871.1 CAL25872.1 CAL25874.1 CAL25875.1 CAL25876.1 CAL25877.1 CAR93019.1 CAR93020.1 CAR93021.1 CAR93022.1 CAR93023.1 CAR93024.1 CAR93025.1 CAR93026.1 CAR93027.1 CAR93028.1 CAR93029.1 CAR93030.1 AY190941 CH954179 AAO01015.1 EDV56742.1 AM293967 AM293971 CAL25869.1 CAL25873.1 KB630005 KB630385 ERL83336.1 ERL83578.1 CCAG010002248 GBHO01021894 GBRD01003021 JAG21710.1 JAG62800.1 EDW60223.1 KQ971327 EEZ99614.1 CP012524 ALC41772.1 KQ414699 KOC63456.1 PNF37500.1 FN546768 CBE67067.1 KQ435724 KOX78353.1 CM002911 KMY95956.1 AM293964 CAL25866.1 CM000362 EDX08308.1 CH480824 EDW56886.1 GAHY01000469 JAA77041.1 ACPB03014062 AJWK01023760 CVRI01000001 CRK86224.1 GEBQ01026283 JAT13694.1 BABH01025880 KQ434846 KZC08368.1 GFTR01006131 JAW10295.1 EDV98355.1 KZ288386 PBC26487.1 KK855555 PTY23106.1 KQ978023 KYM98067.1 GL888439 EGI60841.1 EDW60222.2
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000075886 UP000192223 UP000075900 UP000075881 UP000030765 UP000076408 UP000007062 UP000075883 UP000075901 UP000075884 UP000076407 UP000075903 UP000075882 UP000075840 UP000002320 UP000000673 UP000075880 UP000075885 UP000069272 UP000069940 UP000249989 UP000075920 UP000008820 UP000075902 UP000095300 UP000001819 UP000008744 UP000268350 UP000007798 UP000245037 UP000092462 UP000007801 UP000009192 UP000008792 UP000092443 UP000001070 UP000192221 UP000092445 UP000019118 UP000037069 UP000002282 UP000235965 UP000000803 UP000008711 UP000030742 UP000092444 UP000007266 UP000095301 UP000092553 UP000053825 UP000053105 UP000000304 UP000001292 UP000015103 UP000092461 UP000183832 UP000076502 UP000242457 UP000005203 UP000078542 UP000007755
UP000037510 UP000075886 UP000192223 UP000075900 UP000075881 UP000030765 UP000076408 UP000007062 UP000075883 UP000075901 UP000075884 UP000076407 UP000075903 UP000075882 UP000075840 UP000002320 UP000000673 UP000075880 UP000075885 UP000069272 UP000069940 UP000249989 UP000075920 UP000008820 UP000075902 UP000095300 UP000001819 UP000008744 UP000268350 UP000007798 UP000245037 UP000092462 UP000007801 UP000009192 UP000008792 UP000092443 UP000001070 UP000192221 UP000092445 UP000019118 UP000037069 UP000002282 UP000235965 UP000000803 UP000008711 UP000030742 UP000092444 UP000007266 UP000095301 UP000092553 UP000053825 UP000053105 UP000000304 UP000001292 UP000015103 UP000092461 UP000183832 UP000076502 UP000242457 UP000005203 UP000078542 UP000007755
Interpro
Gene 3D
ProteinModelPortal
H9JQY1
A0A2H1W3Z6
A0A2A4J1A4
A0A3S2NCB3
A0A0N1IPW0
A0A194PWM4
+ More
A0A212FDQ3 A0A2W1C0N9 A0A0L7LLC7 A0A2H1W8M1 A0A0N1II92 A0A336LSN8 A0A194QAF5 A0A182QK68 A0A212F0U9 A0A1W4WGQ3 A0A182RME7 A0A182JQ50 A0A084VY33 A0A182Y3G4 Q7Q482 A0A182MPW5 A0A182SLW7 A0A182MYZ6 A0A182XL58 A0A182V0R4 A0A182L0X1 A0A182ICG6 A0A1Q3FTE8 B0X6Z4 W5J667 A0A182J7Q2 A0A182PCP8 A0A182F4B3 A0A182H7Y6 A0A1Y1N046 A0A182W6P8 A0A182GIL1 A0A2A4J8U6 Q0IFA2 B0X6Z5 A0A1B6GWJ6 A0A182TRF2 A0A1B6CBT4 A0A1I8Q999 Q291Y4 B4GA29 A0A3B0JE42 A0A034W5F2 B4MNL2 W8BMQ2 A0A2P8YAE4 A0A0K8WAA7 A0A1B0D9Q4 B3MF30 B4KSL7 B4LPB5 D1GYC6 A0A1A9XJY6 B4JWL6 A0A1W4VRL2 A0A1B0AJC5 N6TKH4 A0A0L0CD00 B4P924 A0A2J7R9K0 D1GYC9 Q9W1V2 Q8I1F3 A0AMS9 U4TYH0 A0A1B0GCZ6 A0A0A9XWS8 B4LPB4 D6WGQ5 A0A1I8NF91 A0A0M4EH02 A0A0L7QY25 A0A2J7R9H9 D1GZ29 A0A0M9A8X8 A0A0J9RJE3 A0AMS6 B4QI54 B4I8G1 R4FQC8 T1IED1 A0A1B0CQM3 A0A1J1HE31 A0A1B6KR36 H9J9U6 A0A154PAW6 A0A224XCZ4 B4JWL7 A0A2A3E430 A0A087ZQP0 A0A2R7WU91 A0A195CB88 F4WY84 B4LPB3
A0A212FDQ3 A0A2W1C0N9 A0A0L7LLC7 A0A2H1W8M1 A0A0N1II92 A0A336LSN8 A0A194QAF5 A0A182QK68 A0A212F0U9 A0A1W4WGQ3 A0A182RME7 A0A182JQ50 A0A084VY33 A0A182Y3G4 Q7Q482 A0A182MPW5 A0A182SLW7 A0A182MYZ6 A0A182XL58 A0A182V0R4 A0A182L0X1 A0A182ICG6 A0A1Q3FTE8 B0X6Z4 W5J667 A0A182J7Q2 A0A182PCP8 A0A182F4B3 A0A182H7Y6 A0A1Y1N046 A0A182W6P8 A0A182GIL1 A0A2A4J8U6 Q0IFA2 B0X6Z5 A0A1B6GWJ6 A0A182TRF2 A0A1B6CBT4 A0A1I8Q999 Q291Y4 B4GA29 A0A3B0JE42 A0A034W5F2 B4MNL2 W8BMQ2 A0A2P8YAE4 A0A0K8WAA7 A0A1B0D9Q4 B3MF30 B4KSL7 B4LPB5 D1GYC6 A0A1A9XJY6 B4JWL6 A0A1W4VRL2 A0A1B0AJC5 N6TKH4 A0A0L0CD00 B4P924 A0A2J7R9K0 D1GYC9 Q9W1V2 Q8I1F3 A0AMS9 U4TYH0 A0A1B0GCZ6 A0A0A9XWS8 B4LPB4 D6WGQ5 A0A1I8NF91 A0A0M4EH02 A0A0L7QY25 A0A2J7R9H9 D1GZ29 A0A0M9A8X8 A0A0J9RJE3 A0AMS6 B4QI54 B4I8G1 R4FQC8 T1IED1 A0A1B0CQM3 A0A1J1HE31 A0A1B6KR36 H9J9U6 A0A154PAW6 A0A224XCZ4 B4JWL7 A0A2A3E430 A0A087ZQP0 A0A2R7WU91 A0A195CB88 F4WY84 B4LPB3
Ontologies
GO
PANTHER
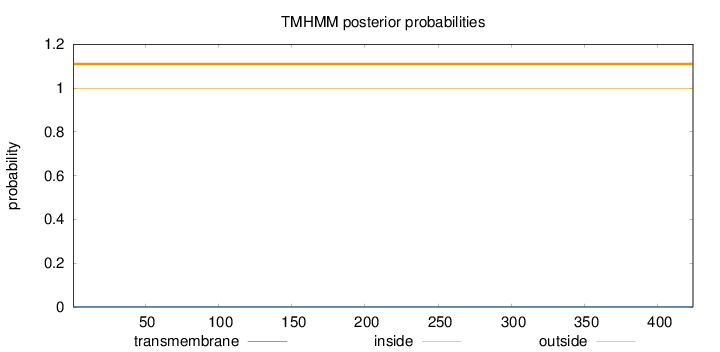
Topology
Length:
424
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00350
outside
1 - 424
Population Genetic Test Statistics
Pi
225.596507
Theta
183.702412
Tajima's D
0.557409
CLR
1.508533
CSRT
0.534923253837308
Interpretation
Uncertain
