Gene
KWMTBOMO06401 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011936
Annotation
H+_transporting_ATP_synthase_subunit_d_[Bombyx_mori]
Full name
ATP synthase subunit d, mitochondrial
Location in the cell
Cytoplasmic Reliability : 1.221 Mitochondrial Reliability : 1.055
Sequence
CDS
ATGGCGAAAAGAATATCACAAAGTGCCGTAAACTGGGCTGCTCTCGCTGAGAGAGTTCCCGCCGAACAAAAGGCGCACCTTGCTGCTTTCAAAATAAAATCCGATAATTATCTCCGAAGGGTACTAGCCAATCCACCTGAACCGCCCAAGATTAACTGGGCTGTGTACAAACAGGCTGTACCTATTCCTGGAATGGTAGACACATTCCAGAAGCAATATGAAGCTCTAAAAATTCCATACCCAGCTGACACTCAGACGGCACTTGTAGAATCGCAGTGGAATCAAGTCAAGAATGCCATCGACGCGTTTATCCAAGAGTCCAATGCCAACATTGCATCCTACCAAAAAGAAATCAATGCAACCAAGGCCCTACTGCCGTATGACCAGATGACCATGGAAGACTTCTATGATGCCCATCCTGACCTGGCCCTTGATCCCATCAAGAAGCCAACCTTCTGGCCACACACTCCGGAAGAGCAGCTCGACTATGTCGACCCAGAGAAACAAGCTCAGCCTACTACAGCTGCAGCTGCACACTAA
Protein
MAKRISQSAVNWAALAERVPAEQKAHLAAFKIKSDNYLRRVLANPPEPPKINWAVYKQAVPIPGMVDTFQKQYEALKIPYPADTQTALVESQWNQVKNAIDAFIQESNANIASYQKEINATKALLPYDQMTMEDFYDAHPDLALDPIKKPTFWPHTPEEQLDYVDPEKQAQPTTAAAAH
Summary
Description
Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core, and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation.
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(0) seems to have nine subunits: a, b, c, d, e, f, g, F6 and 8 (or A6L).
Similarity
Belongs to the ATPase d subunit family.
Keywords
Complete proteome
Hydrogen ion transport
Ion transport
Membrane
Mitochondrion
Mitochondrion inner membrane
Reference proteome
Transport
Feature
chain ATP synthase subunit d, mitochondrial
Uniprot
Q1HPP3
A0A2H1W3Y3
A0A2A4J1Q8
A0A2W1BUC6
A0A194PV08
I4DJX5
+ More
A0A0N0PDJ1 A0A212FDT8 I4DN27 D5LN47 S4PMR7 A0A0L7LM20 A0A3G1T1F9 A0A0L0CLH8 A0A1I8Q4G9 A0A1B0CRW0 T1P7L3 A0A1L8EER5 A0A0K8TEV4 A0A0A9WLZ3 Q1HR21 A0A182N288 A0A182UHJ8 A0A182URL0 A0A182X5Q0 B0WW90 A0A1L8E025 A0A182MP73 A0A182RW42 A0A182KL05 Q7QHC8 A0A2M4A6H1 A0A182FW37 A0A2M4C1B7 A0A182QWM6 A0A034WQQ3 A0A0C9R8N2 D3TN18 A0A2M4A631 A0A069DPS4 A0A0P4VTX2 R4FKB2 A0A182JWZ9 W8BZX3 A0A1B0BJQ8 A0A1Q3FKP2 A0A0K8VFS3 A0A182HW97 A0A2M3YZW3 T1DN10 W5JJY9 B3M1P3 A0A0K8TNX3 A0A182YB01 B4KBB6 A0A2P8XM36 A0A182WDL3 A0A182JHE0 A0A0A1WV14 U5EWQ5 A0A0A1XPL2 A0A0B4LHL7 Q24251 A0A0M4F425 A0A023EHZ0 Q6XIZ3 A0A1A9X1A0 T1E341 B3P374 A0A084WM03 A0A2J7QB91 A0A1B0G0R6 B5DTE3 A0A224XZ38 B4JSF7 A0A067R370 B4MB46 B4ILU9 B4G669 A0A336MJV1 A0A3B0KM59 A0A1W4UB64 N6TZ51 A0A182NZP8 J3JUX8 A0A1J1HWE5 H9J601 Q2XYK8 B4NHY2 Q2XYK7 D6WH57 Q2XYL0 A0A2R7VXW2 Q2XYL1 A0A023F7P5 A0A1B6DLU4 B4QTH4 A0A1W4W3M6 A0A1B0G6D6 A0A232ETU0
A0A0N0PDJ1 A0A212FDT8 I4DN27 D5LN47 S4PMR7 A0A0L7LM20 A0A3G1T1F9 A0A0L0CLH8 A0A1I8Q4G9 A0A1B0CRW0 T1P7L3 A0A1L8EER5 A0A0K8TEV4 A0A0A9WLZ3 Q1HR21 A0A182N288 A0A182UHJ8 A0A182URL0 A0A182X5Q0 B0WW90 A0A1L8E025 A0A182MP73 A0A182RW42 A0A182KL05 Q7QHC8 A0A2M4A6H1 A0A182FW37 A0A2M4C1B7 A0A182QWM6 A0A034WQQ3 A0A0C9R8N2 D3TN18 A0A2M4A631 A0A069DPS4 A0A0P4VTX2 R4FKB2 A0A182JWZ9 W8BZX3 A0A1B0BJQ8 A0A1Q3FKP2 A0A0K8VFS3 A0A182HW97 A0A2M3YZW3 T1DN10 W5JJY9 B3M1P3 A0A0K8TNX3 A0A182YB01 B4KBB6 A0A2P8XM36 A0A182WDL3 A0A182JHE0 A0A0A1WV14 U5EWQ5 A0A0A1XPL2 A0A0B4LHL7 Q24251 A0A0M4F425 A0A023EHZ0 Q6XIZ3 A0A1A9X1A0 T1E341 B3P374 A0A084WM03 A0A2J7QB91 A0A1B0G0R6 B5DTE3 A0A224XZ38 B4JSF7 A0A067R370 B4MB46 B4ILU9 B4G669 A0A336MJV1 A0A3B0KM59 A0A1W4UB64 N6TZ51 A0A182NZP8 J3JUX8 A0A1J1HWE5 H9J601 Q2XYK8 B4NHY2 Q2XYK7 D6WH57 Q2XYL0 A0A2R7VXW2 Q2XYL1 A0A023F7P5 A0A1B6DLU4 B4QTH4 A0A1W4W3M6 A0A1B0G6D6 A0A232ETU0
Pubmed
19121390
28756777
26354079
22651552
22118469
23622113
+ More
26227816 26108605 25315136 26823975 25401762 17204158 17510324 20966253 12364791 14747013 17210077 25348373 20353571 26334808 27129103 24495485 20920257 23761445 17994087 18057021 26369729 25244985 29403074 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10071211 12537569 16120803 24945155 14525923 17550304 24330624 24438588 15632085 24845553 23537049 22516182 18362917 19820115 25474469 28648823
26227816 26108605 25315136 26823975 25401762 17204158 17510324 20966253 12364791 14747013 17210077 25348373 20353571 26334808 27129103 24495485 20920257 23761445 17994087 18057021 26369729 25244985 29403074 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10071211 12537569 16120803 24945155 14525923 17550304 24330624 24438588 15632085 24845553 23537049 22516182 18362917 19820115 25474469 28648823
EMBL
BABH01028570
DQ443359
ABF51448.1
ODYU01006180
SOQ47809.1
NWSH01004140
+ More
PCG65454.1 KZ149897 PZC78682.1 KQ459590 KPI97157.1 AK401593 BAM18215.1 KQ460210 KPJ16587.1 AGBW02008998 OWR51902.1 AK402695 BAM19317.1 GU947650 JN410822 ADF43048.1 AEP25398.1 GAIX01003415 JAA89145.1 JTDY01000680 KOB76246.1 MG992383 AXY94821.1 JRES01000321 KNC32299.1 AJWK01025352 KA644576 AFP59205.1 GFDG01001608 JAV17191.1 GBRD01001807 GDHC01001054 JAG64014.1 JAQ17575.1 GBHO01034142 JAG09462.1 DQ440273 CH477304 ABF18306.1 EAT44176.1 DS232140 EDS35922.1 GFDF01001976 JAV12108.1 AXCM01004806 AAAB01008816 EAA05107.3 GGFK01003034 MBW36355.1 GGFJ01009952 MBW59093.1 AXCN02001731 GAKP01001983 JAC56969.1 GBYB01009207 JAG78974.1 EZ422820 ADD19096.1 GGFK01002943 MBW36264.1 GBGD01003198 JAC85691.1 GDKW01001150 JAI55445.1 ACPB03001919 GAHY01001933 JAA75577.1 GAMC01004287 JAC02269.1 JXJN01015487 GFDL01006999 JAV28046.1 GDHF01014555 JAI37759.1 APCN01001478 GGFM01001052 MBW21803.1 GAMD01003260 JAA98330.1 ADMH02000909 GGFL01004481 ETN64697.1 MBW68659.1 CH902617 EDV44373.1 KPU80816.1 GDAI01001534 JAI16069.1 CH933806 EDW16844.2 PYGN01001739 PSN33066.1 GBXI01011590 JAD02702.1 GANO01001313 JAB58558.1 GBXI01001420 JAD12872.1 AE014297 AHN57400.1 X99667 AY071751 BT088850 DQ138650 CP012526 ALC46070.1 GAPW01004906 JAC08692.1 AY231686 CM000160 AAR09709.1 EDW95985.1 KRK02824.1 GALA01000715 JAA94137.1 CH954181 EDV48526.1 ATLV01024337 KE525351 KFB51247.1 NEVH01016301 PNF25858.1 CCAG010021817 CH475858 EDY71151.1 GFTR01002903 JAW13523.1 CH916373 EDV94697.1 KK852729 KDR17560.1 CH940656 EDW58317.1 KRF78225.1 CH480892 EDW55425.1 CH479179 EDW23828.1 UFQS01001170 UFQS01002160 UFQT01001170 UFQT01002160 SSX09357.1 SSX13389.1 SSX29259.1 OUUW01000013 SPP87659.1 APGK01049756 KB741156 KB631941 ENN73621.1 ERL87374.1 BT127043 AEE62005.1 CVRI01000021 CRK91684.1 BABH01029922 DQ138653 DQ138654 ABA86259.1 ABA86260.1 CH964272 EDW83632.1 KRF99843.1 DQ138655 ABA86261.1 KQ971330 EFA01349.1 DQ138652 ABA86258.1 KK854156 PTY12302.1 DQ138651 ABA86257.1 GBBI01001459 JAC17253.1 GEDC01010653 JAS26645.1 CM000364 EDX12376.1 CCAG010011188 NNAY01002225 OXU21773.1
PCG65454.1 KZ149897 PZC78682.1 KQ459590 KPI97157.1 AK401593 BAM18215.1 KQ460210 KPJ16587.1 AGBW02008998 OWR51902.1 AK402695 BAM19317.1 GU947650 JN410822 ADF43048.1 AEP25398.1 GAIX01003415 JAA89145.1 JTDY01000680 KOB76246.1 MG992383 AXY94821.1 JRES01000321 KNC32299.1 AJWK01025352 KA644576 AFP59205.1 GFDG01001608 JAV17191.1 GBRD01001807 GDHC01001054 JAG64014.1 JAQ17575.1 GBHO01034142 JAG09462.1 DQ440273 CH477304 ABF18306.1 EAT44176.1 DS232140 EDS35922.1 GFDF01001976 JAV12108.1 AXCM01004806 AAAB01008816 EAA05107.3 GGFK01003034 MBW36355.1 GGFJ01009952 MBW59093.1 AXCN02001731 GAKP01001983 JAC56969.1 GBYB01009207 JAG78974.1 EZ422820 ADD19096.1 GGFK01002943 MBW36264.1 GBGD01003198 JAC85691.1 GDKW01001150 JAI55445.1 ACPB03001919 GAHY01001933 JAA75577.1 GAMC01004287 JAC02269.1 JXJN01015487 GFDL01006999 JAV28046.1 GDHF01014555 JAI37759.1 APCN01001478 GGFM01001052 MBW21803.1 GAMD01003260 JAA98330.1 ADMH02000909 GGFL01004481 ETN64697.1 MBW68659.1 CH902617 EDV44373.1 KPU80816.1 GDAI01001534 JAI16069.1 CH933806 EDW16844.2 PYGN01001739 PSN33066.1 GBXI01011590 JAD02702.1 GANO01001313 JAB58558.1 GBXI01001420 JAD12872.1 AE014297 AHN57400.1 X99667 AY071751 BT088850 DQ138650 CP012526 ALC46070.1 GAPW01004906 JAC08692.1 AY231686 CM000160 AAR09709.1 EDW95985.1 KRK02824.1 GALA01000715 JAA94137.1 CH954181 EDV48526.1 ATLV01024337 KE525351 KFB51247.1 NEVH01016301 PNF25858.1 CCAG010021817 CH475858 EDY71151.1 GFTR01002903 JAW13523.1 CH916373 EDV94697.1 KK852729 KDR17560.1 CH940656 EDW58317.1 KRF78225.1 CH480892 EDW55425.1 CH479179 EDW23828.1 UFQS01001170 UFQS01002160 UFQT01001170 UFQT01002160 SSX09357.1 SSX13389.1 SSX29259.1 OUUW01000013 SPP87659.1 APGK01049756 KB741156 KB631941 ENN73621.1 ERL87374.1 BT127043 AEE62005.1 CVRI01000021 CRK91684.1 BABH01029922 DQ138653 DQ138654 ABA86259.1 ABA86260.1 CH964272 EDW83632.1 KRF99843.1 DQ138655 ABA86261.1 KQ971330 EFA01349.1 DQ138652 ABA86258.1 KK854156 PTY12302.1 DQ138651 ABA86257.1 GBBI01001459 JAC17253.1 GEDC01010653 JAS26645.1 CM000364 EDX12376.1 CCAG010011188 NNAY01002225 OXU21773.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000037069 UP000095300 UP000092461 UP000095301 UP000008820 UP000075884 UP000075902 UP000075903 UP000076407 UP000002320 UP000075883 UP000075900 UP000075882 UP000007062 UP000069272 UP000075886 UP000015103 UP000075881 UP000092460 UP000075840 UP000000673 UP000007801 UP000076408 UP000009192 UP000245037 UP000075920 UP000075880 UP000000803 UP000092553 UP000002282 UP000091820 UP000008711 UP000030765 UP000235965 UP000092444 UP000001819 UP000001070 UP000027135 UP000008792 UP000001292 UP000008744 UP000268350 UP000192221 UP000019118 UP000030742 UP000075885 UP000183832 UP000007798 UP000007266 UP000000304 UP000192223 UP000215335
UP000037069 UP000095300 UP000092461 UP000095301 UP000008820 UP000075884 UP000075902 UP000075903 UP000076407 UP000002320 UP000075883 UP000075900 UP000075882 UP000007062 UP000069272 UP000075886 UP000015103 UP000075881 UP000092460 UP000075840 UP000000673 UP000007801 UP000076408 UP000009192 UP000245037 UP000075920 UP000075880 UP000000803 UP000092553 UP000002282 UP000091820 UP000008711 UP000030765 UP000235965 UP000092444 UP000001819 UP000001070 UP000027135 UP000008792 UP000001292 UP000008744 UP000268350 UP000192221 UP000019118 UP000030742 UP000075885 UP000183832 UP000007798 UP000007266 UP000000304 UP000192223 UP000215335
Interpro
Gene 3D
CDD
ProteinModelPortal
Q1HPP3
A0A2H1W3Y3
A0A2A4J1Q8
A0A2W1BUC6
A0A194PV08
I4DJX5
+ More
A0A0N0PDJ1 A0A212FDT8 I4DN27 D5LN47 S4PMR7 A0A0L7LM20 A0A3G1T1F9 A0A0L0CLH8 A0A1I8Q4G9 A0A1B0CRW0 T1P7L3 A0A1L8EER5 A0A0K8TEV4 A0A0A9WLZ3 Q1HR21 A0A182N288 A0A182UHJ8 A0A182URL0 A0A182X5Q0 B0WW90 A0A1L8E025 A0A182MP73 A0A182RW42 A0A182KL05 Q7QHC8 A0A2M4A6H1 A0A182FW37 A0A2M4C1B7 A0A182QWM6 A0A034WQQ3 A0A0C9R8N2 D3TN18 A0A2M4A631 A0A069DPS4 A0A0P4VTX2 R4FKB2 A0A182JWZ9 W8BZX3 A0A1B0BJQ8 A0A1Q3FKP2 A0A0K8VFS3 A0A182HW97 A0A2M3YZW3 T1DN10 W5JJY9 B3M1P3 A0A0K8TNX3 A0A182YB01 B4KBB6 A0A2P8XM36 A0A182WDL3 A0A182JHE0 A0A0A1WV14 U5EWQ5 A0A0A1XPL2 A0A0B4LHL7 Q24251 A0A0M4F425 A0A023EHZ0 Q6XIZ3 A0A1A9X1A0 T1E341 B3P374 A0A084WM03 A0A2J7QB91 A0A1B0G0R6 B5DTE3 A0A224XZ38 B4JSF7 A0A067R370 B4MB46 B4ILU9 B4G669 A0A336MJV1 A0A3B0KM59 A0A1W4UB64 N6TZ51 A0A182NZP8 J3JUX8 A0A1J1HWE5 H9J601 Q2XYK8 B4NHY2 Q2XYK7 D6WH57 Q2XYL0 A0A2R7VXW2 Q2XYL1 A0A023F7P5 A0A1B6DLU4 B4QTH4 A0A1W4W3M6 A0A1B0G6D6 A0A232ETU0
A0A0N0PDJ1 A0A212FDT8 I4DN27 D5LN47 S4PMR7 A0A0L7LM20 A0A3G1T1F9 A0A0L0CLH8 A0A1I8Q4G9 A0A1B0CRW0 T1P7L3 A0A1L8EER5 A0A0K8TEV4 A0A0A9WLZ3 Q1HR21 A0A182N288 A0A182UHJ8 A0A182URL0 A0A182X5Q0 B0WW90 A0A1L8E025 A0A182MP73 A0A182RW42 A0A182KL05 Q7QHC8 A0A2M4A6H1 A0A182FW37 A0A2M4C1B7 A0A182QWM6 A0A034WQQ3 A0A0C9R8N2 D3TN18 A0A2M4A631 A0A069DPS4 A0A0P4VTX2 R4FKB2 A0A182JWZ9 W8BZX3 A0A1B0BJQ8 A0A1Q3FKP2 A0A0K8VFS3 A0A182HW97 A0A2M3YZW3 T1DN10 W5JJY9 B3M1P3 A0A0K8TNX3 A0A182YB01 B4KBB6 A0A2P8XM36 A0A182WDL3 A0A182JHE0 A0A0A1WV14 U5EWQ5 A0A0A1XPL2 A0A0B4LHL7 Q24251 A0A0M4F425 A0A023EHZ0 Q6XIZ3 A0A1A9X1A0 T1E341 B3P374 A0A084WM03 A0A2J7QB91 A0A1B0G0R6 B5DTE3 A0A224XZ38 B4JSF7 A0A067R370 B4MB46 B4ILU9 B4G669 A0A336MJV1 A0A3B0KM59 A0A1W4UB64 N6TZ51 A0A182NZP8 J3JUX8 A0A1J1HWE5 H9J601 Q2XYK8 B4NHY2 Q2XYK7 D6WH57 Q2XYL0 A0A2R7VXW2 Q2XYL1 A0A023F7P5 A0A1B6DLU4 B4QTH4 A0A1W4W3M6 A0A1B0G6D6 A0A232ETU0
PDB
2CLY
E-value=6.67502e-23,
Score=261
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
Mitochondrion
Mitochondrion
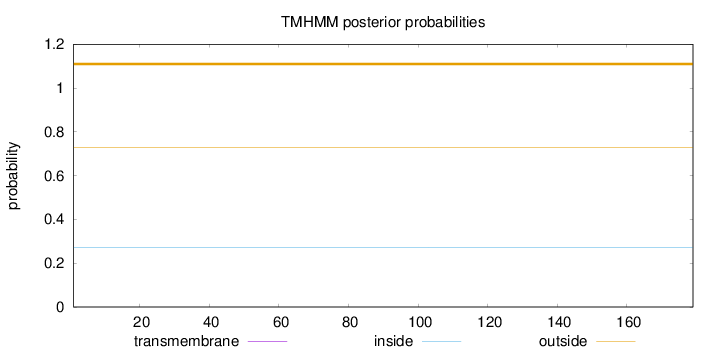
Length:
179
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.27203
outside
1 - 179
Population Genetic Test Statistics
Pi
25.48192
Theta
151.874194
Tajima's D
-0.553458
CLR
23.296078
CSRT
0.232738363081846
Interpretation
Uncertain
