Gene
KWMTBOMO06400
Pre Gene Modal
BGIBMGA011935
Annotation
PREDICTED:_protein_trapped_in_endoderm-1_isoform_X2_[Papilio_xuthus]
Full name
Protein trapped in endoderm-1
Location in the cell
PlasmaMembrane Reliability : 4.996
Sequence
CDS
ATGTCTTCTATCGATAAAATGTATAATTTTACGTTGAGCGACGCGCAAAATGGGACATTTTATCCCACTGAGGCCACGATCTTGGCCGCTGTCTGTGCGAGCGTATTCAGCGTTGTTGGAGTAGTTGGTAACTTAGTAACCGCTGTAGCTCTACTCATGCATCCGAAGCTAAGAGGCCACGTGACCACAATGTTCGTGCTCTCCCTCTGCCTCTCCGATCTACTCTTCTGCGCCATCAACCTGCCTCTAACCGCCAATAGGTTCATCAAGCAACACTGGAGTCTCGGGCCTCAGCTGTGTCAACTTTTCGCGTTCATTTTTTATGGAAATGAAGCCGTGTCACTATTGTCCATGGTGGCTATCACTATTAACAGATACACATTGATAGCATACTACGACATGTACGCGCAAATCTATACGACCACGAAGATATGGATACAATTGCTCCTCATCTGGATCGTGTCTTTCGGATTCATGATTCCCCCCCTGCTCGGCATCTGGGGTCAACTAGGGTTGGAGAAGAACACGTTTTCGTGCACGATATTACCGAAGAACGGGCAGTCGCCGAAAAAAGCGTTGTTCGTTTTCGGCTTCGCGTTGCCCTGCGTCGCGATCATCGTCTCGTACTCGTGCATCTATTGGCGCGTCAGACAGAGCAAACGGAAACTTGAAGGACACGGTAAACTCAGCGGCCAGACTGCGAAAGAGAAAGAAGAAGACTCTAGACTGACGAGGCTGATGCTGACCATCTTCGTGTGCTTCCTCGCCTGCTTCCTGCCGCTGATGATCATGAACGTGGCCGACGACGACATCACGTACCCCTGGCTCCACATCATCGCCTCGATCCTGGCGTGGGCGTCCAGCGTGGTCAACCCGCTGATCTACGCGGCCACCAACAGGCAGTACCGGGCCGCCTACGCCAACCTCCTCAAGTTCTGCAAGCTCAGCCCCGCCTCCCGAAGAACGACGTACGGCAGTCGAACCATGGCGCAGTCCTCCAACTGA
Protein
MSSIDKMYNFTLSDAQNGTFYPTEATILAAVCASVFSVVGVVGNLVTAVALLMHPKLRGHVTTMFVLSLCLSDLLFCAINLPLTANRFIKQHWSLGPQLCQLFAFIFYGNEAVSLLSMVAITINRYTLIAYYDMYAQIYTTTKIWIQLLLIWIVSFGFMIPPLLGIWGQLGLEKNTFSCTILPKNGQSPKKALFVFGFALPCVAIIVSYSCIYWRVRQSKRKLEGHGKLSGQTAKEKEEDSRLTRLMLTIFVCFLACFLPLMIMNVADDDITYPWLHIIASILAWASSVVNPLIYAATNRQYRAAYANLLKFCKLSPASRRTTYGSRTMAQSSN
Summary
Description
Essential for the first active step of germ cell migration: transepithelial migration of germ cells through the posterior midgut (PMG) epithelium.
Miscellaneous
Overexpression of the Tre1 gene restores the taste sensitivity to trehalose in a Tre1 mutant (PubMed:10884225). This experiment cannot be explained given that other authors demonstrate that trehalose sensitivity maps to the adjacent gene, Gr5a (PubMed:14691551).
Similarity
Belongs to the G-protein coupled receptor 1 family.
Keywords
Cell membrane
Complete proteome
G-protein coupled receptor
Glycoprotein
Membrane
Phosphoprotein
Polymorphism
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain Protein trapped in endoderm-1
Uniprot
A0A2W1C3E6
A0A194PUS2
A0A3S2PCC8
A0A0N0PDU5
A0A140B7Z4
A0A212FDT6
+ More
A0A3Q8HDG4 S4NXM5 A0A0L7LL27 D6W7A8 A0A067R423 N6T220 A0A310S3H6 A0A2P8ZH70 A0A088AH57 A0A1Y1L9X6 E9IEH8 A0A195BMR5 A0A158P001 M3UZE9 A0A0E3U2L3 A0A1B0G8C7 A0A1A9Z5T9 A0A195E0Z0 A0A1A9VBM4 A0A0T6B087 A0A195CQI6 A0A1A9XXS6 A0A1B0APR5 A0A151X3T0 F4X3L3 E2ACX8 A0A195F1W8 A0A026VWC8 A0A3L8DVX9 A0A154NYY3 A0A0C9RMU3 A0A0L7RH13 A0A0M8ZZ76 E2BB10 A0A1I8PP66 W8BTX2 A0A232ER08 K7IYK6 A0A0A1XHY2 A0A1I8MNM5 A0A0K8WGU9 A0A0M3QZR8 E9HQ17 B4I0L7 A0A3B0KW52 M9NGQ9 Q9NDM2 A0A2P2IDF3 D2CFN9 C6SUV0 B4L639 A0A0P6EHZ1 B3NV70 B4PZC6 A0A0P6I2T5 A0A1W4VFR0 B4R509 A0A0P6B268 A0A0P6GGD5 X2JIG6 B3MRX8 A0A0K2TB03 A0A0K2TAV5 A0A0P5LQ63 A0A1Q3EUA3 B0W7R2 A0A0N8D974 B4MSG7 A0A1B0CGF8 A0A1B0DPT3 B4M809 Q29I11 B4H4G1 A0A0Q9W6D3 B4JKJ2 T1DIC6 A0A0P5SSI3 A0A1I8PP95 A0A087U3N9 Q17KN3 A0A1D2NGM4 A0A2P8XU17 A0A1W4WBL3 A0A061QFZ2 A0A1W4WBM3 A0A3S3P8F5
A0A3Q8HDG4 S4NXM5 A0A0L7LL27 D6W7A8 A0A067R423 N6T220 A0A310S3H6 A0A2P8ZH70 A0A088AH57 A0A1Y1L9X6 E9IEH8 A0A195BMR5 A0A158P001 M3UZE9 A0A0E3U2L3 A0A1B0G8C7 A0A1A9Z5T9 A0A195E0Z0 A0A1A9VBM4 A0A0T6B087 A0A195CQI6 A0A1A9XXS6 A0A1B0APR5 A0A151X3T0 F4X3L3 E2ACX8 A0A195F1W8 A0A026VWC8 A0A3L8DVX9 A0A154NYY3 A0A0C9RMU3 A0A0L7RH13 A0A0M8ZZ76 E2BB10 A0A1I8PP66 W8BTX2 A0A232ER08 K7IYK6 A0A0A1XHY2 A0A1I8MNM5 A0A0K8WGU9 A0A0M3QZR8 E9HQ17 B4I0L7 A0A3B0KW52 M9NGQ9 Q9NDM2 A0A2P2IDF3 D2CFN9 C6SUV0 B4L639 A0A0P6EHZ1 B3NV70 B4PZC6 A0A0P6I2T5 A0A1W4VFR0 B4R509 A0A0P6B268 A0A0P6GGD5 X2JIG6 B3MRX8 A0A0K2TB03 A0A0K2TAV5 A0A0P5LQ63 A0A1Q3EUA3 B0W7R2 A0A0N8D974 B4MSG7 A0A1B0CGF8 A0A1B0DPT3 B4M809 Q29I11 B4H4G1 A0A0Q9W6D3 B4JKJ2 T1DIC6 A0A0P5SSI3 A0A1I8PP95 A0A087U3N9 Q17KN3 A0A1D2NGM4 A0A2P8XU17 A0A1W4WBL3 A0A061QFZ2 A0A1W4WBM3 A0A3S3P8F5
Pubmed
28756777
26354079
26930056
22118469
23622113
26227816
+ More
18362917 19820115 24845553 23537049 29403074 28004739 21282665 21347285 23517120 25856077 21719571 20798317 24508170 30249741 24495485 28648823 20075255 25830018 25315136 21292972 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10884225 11566105 12537569 14691551 18327897 17550304 18057021 15632085 24330624 17510324 27289101
18362917 19820115 24845553 23537049 29403074 28004739 21282665 21347285 23517120 25856077 21719571 20798317 24508170 30249741 24495485 28648823 20075255 25830018 25315136 21292972 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10884225 11566105 12537569 14691551 18327897 17550304 18057021 15632085 24330624 17510324 27289101
EMBL
KZ149897
PZC78683.1
KQ459590
KPI97156.1
RSAL01000104
RVE47371.1
+ More
KQ460210 KPJ16588.1 KT860048 ALS03936.1 AGBW02008998 OWR51901.1 MF625599 AXY83426.1 GAIX01014130 JAA78430.1 JTDY01000680 KOB76248.1 KQ971307 EFA11575.1 KK852996 KDR12697.1 APGK01047123 APGK01047124 KB741077 KB632384 ENN74144.1 ERL94278.1 GGOB01000040 MCH29436.1 PYGN01000058 PSN55846.1 GEZM01063405 JAV69110.1 GL762593 EFZ21025.1 KQ976440 KYM86576.1 ADTU01005183 GACR01000044 JAA74417.1 KM251701 AKC58582.1 CCAG010000239 KQ979955 KYN18544.1 LJIG01016349 KRT80867.1 KQ977394 KYN03008.1 JXJN01001549 JXJN01001550 KQ982557 KYQ55086.1 GL888624 EGI58810.1 GL438602 EFN68708.1 KQ981856 KYN34570.1 KK107929 EZA47109.1 QOIP01000003 RLU24614.1 KQ434775 KZC04080.1 GBYB01009685 JAG79452.1 KQ414591 KOC70267.1 KQ435794 KOX74110.1 GL446901 EFN87101.1 GAMC01003914 GAMC01003913 JAC02643.1 NNAY01002687 OXU20771.1 GBXI01003767 JAD10525.1 GDHF01002214 JAI50100.1 CP012528 ALC49893.1 GL732713 EFX66166.1 CH480819 EDW53048.1 OUUW01000025 SPP89631.1 AE014298 AFH07248.1 AB034204 AB042625 AB066613 AB066614 AB066615 AB066616 AB066617 AB066618 AY070980 IACF01006456 LAB72039.1 DQ666846 ABH01174.1 BT088942 ACT76688.1 CH933812 EDW05835.1 GDIQ01076346 LRGB01000359 JAN18391.1 KZS19551.1 CH954180 EDV45918.2 CM000162 EDX01093.1 GDIQ01011374 JAN83363.1 CM000366 EDX17158.1 GDIP01021067 JAM82648.1 GDIQ01053579 JAN41158.1 AHN59372.1 CH902622 EDV34533.1 HACA01005832 CDW23193.1 HACA01005833 CDW23194.1 GDIQ01166778 JAK84947.1 GFDL01016154 JAV18891.1 DS231855 EDS38206.1 GDIP01056455 JAM47260.1 CH963851 EDW75056.1 AJWK01011081 AJVK01008240 AJVK01008241 CH940653 EDW62285.1 KRF80538.1 CH379064 EAL31595.2 CH479209 EDW32712.1 KRF80537.1 CH916370 EDW00095.1 GALA01001001 JAA93851.1 GDIP01135902 JAL67812.1 KK118013 KFM71978.1 CH477222 EAT47271.2 LJIJ01000044 ODN04410.1 PYGN01001348 PSN35490.1 GBFC01000021 JAC59317.1 NCKU01006631 RWS03304.1
KQ460210 KPJ16588.1 KT860048 ALS03936.1 AGBW02008998 OWR51901.1 MF625599 AXY83426.1 GAIX01014130 JAA78430.1 JTDY01000680 KOB76248.1 KQ971307 EFA11575.1 KK852996 KDR12697.1 APGK01047123 APGK01047124 KB741077 KB632384 ENN74144.1 ERL94278.1 GGOB01000040 MCH29436.1 PYGN01000058 PSN55846.1 GEZM01063405 JAV69110.1 GL762593 EFZ21025.1 KQ976440 KYM86576.1 ADTU01005183 GACR01000044 JAA74417.1 KM251701 AKC58582.1 CCAG010000239 KQ979955 KYN18544.1 LJIG01016349 KRT80867.1 KQ977394 KYN03008.1 JXJN01001549 JXJN01001550 KQ982557 KYQ55086.1 GL888624 EGI58810.1 GL438602 EFN68708.1 KQ981856 KYN34570.1 KK107929 EZA47109.1 QOIP01000003 RLU24614.1 KQ434775 KZC04080.1 GBYB01009685 JAG79452.1 KQ414591 KOC70267.1 KQ435794 KOX74110.1 GL446901 EFN87101.1 GAMC01003914 GAMC01003913 JAC02643.1 NNAY01002687 OXU20771.1 GBXI01003767 JAD10525.1 GDHF01002214 JAI50100.1 CP012528 ALC49893.1 GL732713 EFX66166.1 CH480819 EDW53048.1 OUUW01000025 SPP89631.1 AE014298 AFH07248.1 AB034204 AB042625 AB066613 AB066614 AB066615 AB066616 AB066617 AB066618 AY070980 IACF01006456 LAB72039.1 DQ666846 ABH01174.1 BT088942 ACT76688.1 CH933812 EDW05835.1 GDIQ01076346 LRGB01000359 JAN18391.1 KZS19551.1 CH954180 EDV45918.2 CM000162 EDX01093.1 GDIQ01011374 JAN83363.1 CM000366 EDX17158.1 GDIP01021067 JAM82648.1 GDIQ01053579 JAN41158.1 AHN59372.1 CH902622 EDV34533.1 HACA01005832 CDW23193.1 HACA01005833 CDW23194.1 GDIQ01166778 JAK84947.1 GFDL01016154 JAV18891.1 DS231855 EDS38206.1 GDIP01056455 JAM47260.1 CH963851 EDW75056.1 AJWK01011081 AJVK01008240 AJVK01008241 CH940653 EDW62285.1 KRF80538.1 CH379064 EAL31595.2 CH479209 EDW32712.1 KRF80537.1 CH916370 EDW00095.1 GALA01001001 JAA93851.1 GDIP01135902 JAL67812.1 KK118013 KFM71978.1 CH477222 EAT47271.2 LJIJ01000044 ODN04410.1 PYGN01001348 PSN35490.1 GBFC01000021 JAC59317.1 NCKU01006631 RWS03304.1
Proteomes
UP000053268
UP000283053
UP000053240
UP000007151
UP000037510
UP000007266
+ More
UP000027135 UP000019118 UP000030742 UP000245037 UP000005203 UP000078540 UP000005205 UP000092444 UP000092445 UP000078492 UP000078200 UP000078542 UP000092443 UP000092460 UP000075809 UP000007755 UP000000311 UP000078541 UP000053097 UP000279307 UP000076502 UP000053825 UP000053105 UP000008237 UP000095300 UP000215335 UP000002358 UP000095301 UP000092553 UP000000305 UP000001292 UP000268350 UP000000803 UP000009192 UP000076858 UP000008711 UP000002282 UP000192221 UP000000304 UP000007801 UP000002320 UP000007798 UP000092461 UP000092462 UP000008792 UP000001819 UP000008744 UP000001070 UP000054359 UP000008820 UP000094527 UP000192223 UP000285301
UP000027135 UP000019118 UP000030742 UP000245037 UP000005203 UP000078540 UP000005205 UP000092444 UP000092445 UP000078492 UP000078200 UP000078542 UP000092443 UP000092460 UP000075809 UP000007755 UP000000311 UP000078541 UP000053097 UP000279307 UP000076502 UP000053825 UP000053105 UP000008237 UP000095300 UP000215335 UP000002358 UP000095301 UP000092553 UP000000305 UP000001292 UP000268350 UP000000803 UP000009192 UP000076858 UP000008711 UP000002282 UP000192221 UP000000304 UP000007801 UP000002320 UP000007798 UP000092461 UP000092462 UP000008792 UP000001819 UP000008744 UP000001070 UP000054359 UP000008820 UP000094527 UP000192223 UP000285301
Pfam
PF00001 7tm_1
ProteinModelPortal
A0A2W1C3E6
A0A194PUS2
A0A3S2PCC8
A0A0N0PDU5
A0A140B7Z4
A0A212FDT6
+ More
A0A3Q8HDG4 S4NXM5 A0A0L7LL27 D6W7A8 A0A067R423 N6T220 A0A310S3H6 A0A2P8ZH70 A0A088AH57 A0A1Y1L9X6 E9IEH8 A0A195BMR5 A0A158P001 M3UZE9 A0A0E3U2L3 A0A1B0G8C7 A0A1A9Z5T9 A0A195E0Z0 A0A1A9VBM4 A0A0T6B087 A0A195CQI6 A0A1A9XXS6 A0A1B0APR5 A0A151X3T0 F4X3L3 E2ACX8 A0A195F1W8 A0A026VWC8 A0A3L8DVX9 A0A154NYY3 A0A0C9RMU3 A0A0L7RH13 A0A0M8ZZ76 E2BB10 A0A1I8PP66 W8BTX2 A0A232ER08 K7IYK6 A0A0A1XHY2 A0A1I8MNM5 A0A0K8WGU9 A0A0M3QZR8 E9HQ17 B4I0L7 A0A3B0KW52 M9NGQ9 Q9NDM2 A0A2P2IDF3 D2CFN9 C6SUV0 B4L639 A0A0P6EHZ1 B3NV70 B4PZC6 A0A0P6I2T5 A0A1W4VFR0 B4R509 A0A0P6B268 A0A0P6GGD5 X2JIG6 B3MRX8 A0A0K2TB03 A0A0K2TAV5 A0A0P5LQ63 A0A1Q3EUA3 B0W7R2 A0A0N8D974 B4MSG7 A0A1B0CGF8 A0A1B0DPT3 B4M809 Q29I11 B4H4G1 A0A0Q9W6D3 B4JKJ2 T1DIC6 A0A0P5SSI3 A0A1I8PP95 A0A087U3N9 Q17KN3 A0A1D2NGM4 A0A2P8XU17 A0A1W4WBL3 A0A061QFZ2 A0A1W4WBM3 A0A3S3P8F5
A0A3Q8HDG4 S4NXM5 A0A0L7LL27 D6W7A8 A0A067R423 N6T220 A0A310S3H6 A0A2P8ZH70 A0A088AH57 A0A1Y1L9X6 E9IEH8 A0A195BMR5 A0A158P001 M3UZE9 A0A0E3U2L3 A0A1B0G8C7 A0A1A9Z5T9 A0A195E0Z0 A0A1A9VBM4 A0A0T6B087 A0A195CQI6 A0A1A9XXS6 A0A1B0APR5 A0A151X3T0 F4X3L3 E2ACX8 A0A195F1W8 A0A026VWC8 A0A3L8DVX9 A0A154NYY3 A0A0C9RMU3 A0A0L7RH13 A0A0M8ZZ76 E2BB10 A0A1I8PP66 W8BTX2 A0A232ER08 K7IYK6 A0A0A1XHY2 A0A1I8MNM5 A0A0K8WGU9 A0A0M3QZR8 E9HQ17 B4I0L7 A0A3B0KW52 M9NGQ9 Q9NDM2 A0A2P2IDF3 D2CFN9 C6SUV0 B4L639 A0A0P6EHZ1 B3NV70 B4PZC6 A0A0P6I2T5 A0A1W4VFR0 B4R509 A0A0P6B268 A0A0P6GGD5 X2JIG6 B3MRX8 A0A0K2TB03 A0A0K2TAV5 A0A0P5LQ63 A0A1Q3EUA3 B0W7R2 A0A0N8D974 B4MSG7 A0A1B0CGF8 A0A1B0DPT3 B4M809 Q29I11 B4H4G1 A0A0Q9W6D3 B4JKJ2 T1DIC6 A0A0P5SSI3 A0A1I8PP95 A0A087U3N9 Q17KN3 A0A1D2NGM4 A0A2P8XU17 A0A1W4WBL3 A0A061QFZ2 A0A1W4WBM3 A0A3S3P8F5
PDB
5DHH
E-value=2.36653e-14,
Score=191
Ontologies
GO
Topology
Subcellular location
Cell membrane
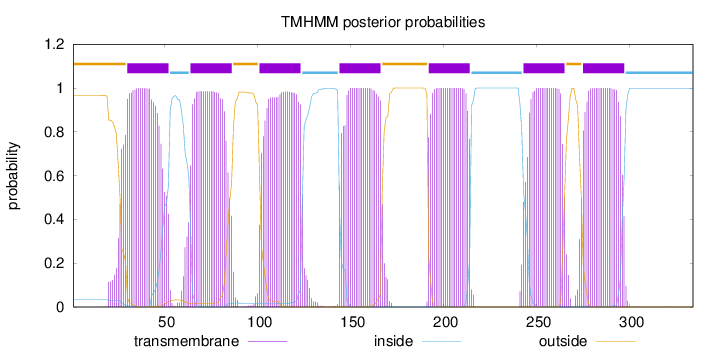
Length:
334
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
157.38341
Exp number, first 60 AAs:
24.29264
Total prob of N-in:
0.03335
POSSIBLE N-term signal
sequence
outside
1 - 29
TMhelix
30 - 52
inside
53 - 63
TMhelix
64 - 86
outside
87 - 100
TMhelix
101 - 123
inside
124 - 143
TMhelix
144 - 166
outside
167 - 191
TMhelix
192 - 214
inside
215 - 242
TMhelix
243 - 265
outside
266 - 274
TMhelix
275 - 297
inside
298 - 334
Population Genetic Test Statistics
Pi
211.010353
Theta
155.889826
Tajima's D
1.122586
CLR
0.430838
CSRT
0.687515624218789
Interpretation
Uncertain
