Pre Gene Modal
BGIBMGA011966
Annotation
CDKAL1-like_protein_[Papilio_xuthus]
Full name
Xylose isomerase
+ More
Threonylcarbamoyladenosine tRNA methylthiotransferase
Threonylcarbamoyladenosine tRNA methylthiotransferase
Alternative Name
CDKAL1-like protein
tRNA-t(6)A37 methylthiotransferase
CDK5 regulatory subunit-associated protein 1-like 1
tRNA-t(6)A37 methylthiotransferase
CDK5 regulatory subunit-associated protein 1-like 1
Location in the cell
Cytoplasmic Reliability : 2.118
Sequence
CDS
ATGCCAGGTGTTTGTGATGAAATTATAGACGACATAGAAGATTTGATATCTTCCCAGGATATAACTCCTAAAGAACGTTATGCTAGTAGGAAGAATGTAAGCGTCCGATCTAAAAAGAGGGAGAAAAAAGACCCCGAACAGATTGAAAAGGTTATATTAGAGAGTGTTGTACCAGGCACTCAAACAATATATGTAAAAACATGGGGCTGTGCTCATAACAACTCAGATTCAGAGTACATGGCCGGACTGCTGGCTGCTAATGGCTACAAATTGACAGAAGACAAATGGGATGCGCAACTTTGGTTGTTGAATTCATGTACTGTAAAGAGCCCGGCTGAGGATCATTTCAAGAATGAAATTGAGCTTGGTCAGAGTCGTGGCATACATGTTGTTGTAGCAGGCTGTGTACCGCAGGGCGCACCAAAAAGTGGCTACCTACATGGACTCAGTATAGTTGGTGTACAACAGATTGACAGAATTGTGGAGGTCGTGGAGGAGACTTTGAAAGGTCACACAGTCCGTCTGTTTGGTCAGAGGAAAACAAACGGTCGTAAAGCGGGTGGGGCTTCACTGCTCCTGCCCAAAGTGCGCAAAAATCCTTTAGTCGAGATCATAGCCGTGAACACGGGCTGCCTGAACCAGTGCACATACTGCAAGACCAAGCACGCCAGAGGAGAGCTGGGCAGCTACCCTCCGGAAGAGATCGTGGAGAGAGCACGACAGTCGTTCACCGAGGGAGTTGTGGAGATATGGCTTACAAGCGAGGATACAGGAACCTACGGTCGTGACATAGGAACATCTCTGCCTGCCCTGCTCGATGCGCTAGTATCAGTGATACCGGACGGCACGAGGCTCCGTCTGGGCATGACCAACCCTCCCTACATCCTGGAACACCTGCCCGCTGTCGCGAGGATCATGAGGCACCCCAGGGTCTACAAGTTTCTCCACGTCCCTGTGCAGTCGGCCAGTGACCAGGTACTGTCCGACATGAAGAGGGAATACACGCGAGCCGACTTCGAACACGTGGTCGACTATCTCAGGAAAGAAGTCCCCGGCATCACGATAGCCACAGACATCATCTGCGGCTTCCCCACTGAGACCGAAGCTGACTTCGAGGAGACCATGCAGCTCTGCAGAAAGTACAAGTTCCCTTCGTTGTTCATCAACCAGTTCTTCCCGAGACCCGGGACGCCGGCTGCCAAGATGACCAGGGTACCTGGTCAAGAAGTAAAGAAACGCACAAAATTACTGTCGGACTTCTTCAGGTCGTACGAACCTTACGGTCACAAGGTGGGGGAGACGCAGGAGGTGCTAGTGACCGACGTGTCGCACGACAAGAACTACTTTGTCGGCCACAACGAGTTCTACGAGCAAGTACTGATACCTAAGGAGGAGAAGTACATGGGGAAATTAGTTACAGTAAAGATTACGAGCGCCAGTAAATTCTCTATGATGGGTCAGCCCATAGACAAACCTAAGATGGCGGGCTTGACGGTGCCTTTGAAGAAAGGAGAGCTTTCCGGTGTCTATGGTTTGAAAAGGGAAAATAAAATACCTATCCCTATACTTGTGCTGTTGATGGTTGTCTTAGCTAGATTTATATGGATGTTTTTTATAGTTTAA
Protein
MPGVCDEIIDDIEDLISSQDITPKERYASRKNVSVRSKKREKKDPEQIEKVILESVVPGTQTIYVKTWGCAHNNSDSEYMAGLLAANGYKLTEDKWDAQLWLLNSCTVKSPAEDHFKNEIELGQSRGIHVVVAGCVPQGAPKSGYLHGLSIVGVQQIDRIVEVVEETLKGHTVRLFGQRKTNGRKAGGASLLLPKVRKNPLVEIIAVNTGCLNQCTYCKTKHARGELGSYPPEEIVERARQSFTEGVVEIWLTSEDTGTYGRDIGTSLPALLDALVSVIPDGTRLRLGMTNPPYILEHLPAVARIMRHPRVYKFLHVPVQSASDQVLSDMKREYTRADFEHVVDYLRKEVPGITIATDIICGFPTETEADFEETMQLCRKYKFPSLFINQFFPRPGTPAAKMTRVPGQEVKKRTKLLSDFFRSYEPYGHKVGETQEVLVTDVSHDKNYFVGHNEFYEQVLIPKEEKYMGKLVTVKITSASKFSMMGQPIDKPKMAGLTVPLKKGELSGVYGLKRENKIPIPILVLLMVVLARFIWMFFIV
Summary
Description
Catalyzes the methylthiolation of N6-threonylcarbamoyladenosine (t(6)A), leading to the formation of 2-methylthio-N6-threonylcarbamoyladenosine (ms(2)t(6)A) at position 37 in tRNAs that read codons beginning with adenine.
Catalytic Activity
D-xylose = D-xylulose
[sulfur carrier]-SH + AH2 + N(6)-L-threonylcarbamoyladenosine(37) in tRNA + 2 S-adenosyl-L-methionine = 2-methylsulfanyl-N(6)-L-threonylcarbamoyladenosine(37) in tRNA + 5'-deoxyadenosine + [sulfur carrier]-H + A + 2 H(+) + L-methionine + S-adenosyl-L-homocysteine
[sulfur carrier]-SH + AH2 + N(6)-L-threonylcarbamoyladenosine(37) in tRNA + 2 S-adenosyl-L-methionine = 2-methylsulfanyl-N(6)-L-threonylcarbamoyladenosine(37) in tRNA + 5'-deoxyadenosine + [sulfur carrier]-H + A + 2 H(+) + L-methionine + S-adenosyl-L-homocysteine
Cofactor
[4Fe-4S] cluster
Similarity
Belongs to the xylose isomerase family.
Belongs to the methylthiotransferase family. CDKAL1 subfamily.
Belongs to the E2F/DP family.
Belongs to the methylthiotransferase family. CDKAL1 subfamily.
Belongs to the E2F/DP family.
Keywords
4Fe-4S
Complete proteome
Iron
Iron-sulfur
Membrane
Metal-binding
Reference proteome
S-adenosyl-L-methionine
Transferase
Transmembrane
Transmembrane helix
tRNA processing
Endoplasmic reticulum
Feature
chain Threonylcarbamoyladenosine tRNA methylthiotransferase
Uniprot
H9JR09
A0A194Q181
A0A2A4JIL5
A0A2H1VNN1
S4PM93
F1C4C0
+ More
A0A2W1BYK8 A0A212FDU8 D6W6D0 A0A151J017 A0A026WQS6 E9ICL2 A0A195FNM0 A0A2J7PJS5 A0A195CVR9 E1ZVZ2 A0A0M8ZXM5 A0A2A3ESV7 A0A0J7L5J6 A0A154P5T8 F4WMT0 A0A1Y1M1F3 A0A0L7QPS3 A0A195BJ44 A0A158NQ43 U4U8D7 E9FR10 B4NQ57 A0A0P5N242 A0A3B0JMC1 A0A0P5VGW0 A0A0P6HHG8 A0A0P5AZR9 A0A3B0JUD9 A0A0P6F6N9 A0A0P4YGR0 B3MG65 A0A0P5VCT9 A0A0P5U398 A0A1B6CSR2 A0A1W4VDB4 A0A1B0CY58 B4LPW9 B4J918 A0A1B0DK13 B3NNR9 A0A0P4WFE8 Q7K4W1 A0A1B6KGQ2 A0A232EI88 B4QAT0 B4HML1 A0A1I8NAH0 A0A1B0DFB9 A0A0P5PKE8 Q16Q27 A0A1I8NZW7 A0A0M4EU02 A7S7F5 A0A0K8TM36 E2BHJ6 A0A1Q3F360 A0A1B6EL38 A0A1B6G2W2 A0A210PNT2 B0W0A5 A0A1A9WNI8 W8BFT4 A0A1B0G4Q9 A0A023F4W3 A0A1A9Y303 A0A1A9ZKW6 V9KMV6 A0A069DUI9 A0A3B3DID5 A0A1S3IFX3 B4KQZ9 A0A1B0C2H0 B4GAQ6 A0A3B3DIE0 A0A3B3C2A1 A0A1A9VKQ5 Q291H5 A0A1L8FSF7 A0A224XM19 A0A0S7FQS5 Q6NS26 T1ICA5 A0A3B4BDX2 A0A091WK03 A0A3B4UQR5 A0A3B4URE2 U3KGF0 A0A3B4UT16 A0A0A1WDX7 A0A3Q2CNL6 A0A1V4JGI9 R0LT49 A0A1A8UAR2 A0A2G8JTL9
A0A2W1BYK8 A0A212FDU8 D6W6D0 A0A151J017 A0A026WQS6 E9ICL2 A0A195FNM0 A0A2J7PJS5 A0A195CVR9 E1ZVZ2 A0A0M8ZXM5 A0A2A3ESV7 A0A0J7L5J6 A0A154P5T8 F4WMT0 A0A1Y1M1F3 A0A0L7QPS3 A0A195BJ44 A0A158NQ43 U4U8D7 E9FR10 B4NQ57 A0A0P5N242 A0A3B0JMC1 A0A0P5VGW0 A0A0P6HHG8 A0A0P5AZR9 A0A3B0JUD9 A0A0P6F6N9 A0A0P4YGR0 B3MG65 A0A0P5VCT9 A0A0P5U398 A0A1B6CSR2 A0A1W4VDB4 A0A1B0CY58 B4LPW9 B4J918 A0A1B0DK13 B3NNR9 A0A0P4WFE8 Q7K4W1 A0A1B6KGQ2 A0A232EI88 B4QAT0 B4HML1 A0A1I8NAH0 A0A1B0DFB9 A0A0P5PKE8 Q16Q27 A0A1I8NZW7 A0A0M4EU02 A7S7F5 A0A0K8TM36 E2BHJ6 A0A1Q3F360 A0A1B6EL38 A0A1B6G2W2 A0A210PNT2 B0W0A5 A0A1A9WNI8 W8BFT4 A0A1B0G4Q9 A0A023F4W3 A0A1A9Y303 A0A1A9ZKW6 V9KMV6 A0A069DUI9 A0A3B3DID5 A0A1S3IFX3 B4KQZ9 A0A1B0C2H0 B4GAQ6 A0A3B3DIE0 A0A3B3C2A1 A0A1A9VKQ5 Q291H5 A0A1L8FSF7 A0A224XM19 A0A0S7FQS5 Q6NS26 T1ICA5 A0A3B4BDX2 A0A091WK03 A0A3B4UQR5 A0A3B4URE2 U3KGF0 A0A3B4UT16 A0A0A1WDX7 A0A3Q2CNL6 A0A1V4JGI9 R0LT49 A0A1A8UAR2 A0A2G8JTL9
EC Number
5.3.1.5
2.8.4.5
2.8.4.5
Pubmed
19121390
26354079
23622113
28756777
22118469
18362917
+ More
19820115 24508170 30249741 21282665 20798317 21719571 28004739 21347285 23537049 21292972 17994087 10731132 12537572 12537569 28648823 22936249 25315136 17510324 17615350 26369729 28812685 24495485 25474469 24402279 26334808 29451363 15632085 27762356 25463417 25830018 29023486
19820115 24508170 30249741 21282665 20798317 21719571 28004739 21347285 23537049 21292972 17994087 10731132 12537572 12537569 28648823 22936249 25315136 17510324 17615350 26369729 28812685 24495485 25474469 24402279 26334808 29451363 15632085 27762356 25463417 25830018 29023486
EMBL
BABH01028575
KQ459590
KPI97155.1
NWSH01001334
PCG71649.1
ODYU01003295
+ More
SOQ41864.1 GAIX01003695 JAA88865.1 HQ199330 ADX30521.1 KZ149897 PZC78684.1 AGBW02008998 OWR51900.1 KQ971319 EFA11366.1 KQ980658 KYN14779.1 KK107148 QOIP01000009 EZA57454.1 RLU18877.1 GL762326 EFZ21687.1 KQ981382 KYN42150.1 NEVH01024946 PNF16584.1 KQ977279 KYN04254.1 GL434696 EFN74642.1 KQ435802 KOX73195.1 KZ288186 PBC34863.1 LBMM01000671 KMQ97823.1 KQ434822 KZC07211.1 GL888218 EGI64557.1 GEZM01042817 JAV79573.1 KQ414805 KOC60637.1 KQ976464 KYM84417.1 ADTU01022953 KB631869 ERL86836.1 GL732523 EFX90271.1 CH964291 EDW86282.1 GDIQ01148434 JAL03292.1 OUUW01000001 SPP74689.1 GDIP01101779 JAM01936.1 GDIQ01028109 JAN66628.1 GDIP01191974 GDIP01049187 LRGB01000248 JAJ31428.1 KZS20098.1 SPP74688.1 GDIQ01053862 JAN40875.1 GDIP01231227 JAI92174.1 CH902619 EDV36760.1 GDIP01101780 JAM01935.1 GDIP01118449 JAL85265.1 GEDC01020923 JAS16375.1 AJWK01035490 CH940648 EDW61309.1 CH916367 EDW01367.1 AJVK01035195 CH954179 EDV55626.1 GDRN01060569 JAI65341.1 AE013599 AY051634 GEBQ01029378 JAT10599.1 NNAY01004341 OXU18031.1 CM000362 CM002911 EDX07481.1 KMY94531.1 CH480816 EDW48279.1 AJVK01058655 GDIQ01132638 JAL19088.1 CH477765 EAT36482.1 CP012524 ALC41087.1 DS469592 EDO40392.1 GDAI01002176 JAI15427.1 GL448301 EFN84849.1 GFDL01013051 JAV21994.1 GECZ01031130 JAS38639.1 GECZ01012989 JAS56780.1 NEDP02005572 OWF38160.1 DS231817 EDS39644.1 GAMC01006390 JAC00166.1 CCAG010010868 GBBI01002678 JAC16034.1 JW866821 AFO99338.1 GBGD01001101 JAC87788.1 CH933808 EDW10355.1 JXJN01024553 CH479181 EDW32008.1 CM000071 CM004477 OCT74519.1 GFTR01006784 JAW09642.1 GBYX01462234 JAO19351.1 BC070521 ACPB03004219 KK735815 KFR15987.1 AGTO01010359 GBXI01017426 JAC96865.1 LSYS01007707 OPJ71323.1 KB742735 EOB04920.1 HADY01006470 HAEJ01003746 SBS44203.1 MRZV01001275 PIK39117.1
SOQ41864.1 GAIX01003695 JAA88865.1 HQ199330 ADX30521.1 KZ149897 PZC78684.1 AGBW02008998 OWR51900.1 KQ971319 EFA11366.1 KQ980658 KYN14779.1 KK107148 QOIP01000009 EZA57454.1 RLU18877.1 GL762326 EFZ21687.1 KQ981382 KYN42150.1 NEVH01024946 PNF16584.1 KQ977279 KYN04254.1 GL434696 EFN74642.1 KQ435802 KOX73195.1 KZ288186 PBC34863.1 LBMM01000671 KMQ97823.1 KQ434822 KZC07211.1 GL888218 EGI64557.1 GEZM01042817 JAV79573.1 KQ414805 KOC60637.1 KQ976464 KYM84417.1 ADTU01022953 KB631869 ERL86836.1 GL732523 EFX90271.1 CH964291 EDW86282.1 GDIQ01148434 JAL03292.1 OUUW01000001 SPP74689.1 GDIP01101779 JAM01936.1 GDIQ01028109 JAN66628.1 GDIP01191974 GDIP01049187 LRGB01000248 JAJ31428.1 KZS20098.1 SPP74688.1 GDIQ01053862 JAN40875.1 GDIP01231227 JAI92174.1 CH902619 EDV36760.1 GDIP01101780 JAM01935.1 GDIP01118449 JAL85265.1 GEDC01020923 JAS16375.1 AJWK01035490 CH940648 EDW61309.1 CH916367 EDW01367.1 AJVK01035195 CH954179 EDV55626.1 GDRN01060569 JAI65341.1 AE013599 AY051634 GEBQ01029378 JAT10599.1 NNAY01004341 OXU18031.1 CM000362 CM002911 EDX07481.1 KMY94531.1 CH480816 EDW48279.1 AJVK01058655 GDIQ01132638 JAL19088.1 CH477765 EAT36482.1 CP012524 ALC41087.1 DS469592 EDO40392.1 GDAI01002176 JAI15427.1 GL448301 EFN84849.1 GFDL01013051 JAV21994.1 GECZ01031130 JAS38639.1 GECZ01012989 JAS56780.1 NEDP02005572 OWF38160.1 DS231817 EDS39644.1 GAMC01006390 JAC00166.1 CCAG010010868 GBBI01002678 JAC16034.1 JW866821 AFO99338.1 GBGD01001101 JAC87788.1 CH933808 EDW10355.1 JXJN01024553 CH479181 EDW32008.1 CM000071 CM004477 OCT74519.1 GFTR01006784 JAW09642.1 GBYX01462234 JAO19351.1 BC070521 ACPB03004219 KK735815 KFR15987.1 AGTO01010359 GBXI01017426 JAC96865.1 LSYS01007707 OPJ71323.1 KB742735 EOB04920.1 HADY01006470 HAEJ01003746 SBS44203.1 MRZV01001275 PIK39117.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000007266
UP000078492
+ More
UP000053097 UP000279307 UP000078541 UP000235965 UP000078542 UP000000311 UP000053105 UP000242457 UP000036403 UP000076502 UP000007755 UP000053825 UP000078540 UP000005205 UP000030742 UP000000305 UP000007798 UP000268350 UP000076858 UP000007801 UP000192221 UP000092461 UP000008792 UP000001070 UP000092462 UP000008711 UP000000803 UP000215335 UP000000304 UP000001292 UP000095301 UP000008820 UP000095300 UP000092553 UP000001593 UP000008237 UP000242188 UP000002320 UP000091820 UP000092444 UP000092443 UP000092445 UP000261560 UP000085678 UP000009192 UP000092460 UP000008744 UP000078200 UP000001819 UP000186698 UP000015103 UP000261520 UP000053605 UP000261420 UP000016665 UP000265020 UP000190648 UP000230750
UP000053097 UP000279307 UP000078541 UP000235965 UP000078542 UP000000311 UP000053105 UP000242457 UP000036403 UP000076502 UP000007755 UP000053825 UP000078540 UP000005205 UP000030742 UP000000305 UP000007798 UP000268350 UP000076858 UP000007801 UP000192221 UP000092461 UP000008792 UP000001070 UP000092462 UP000008711 UP000000803 UP000215335 UP000000304 UP000001292 UP000095301 UP000008820 UP000095300 UP000092553 UP000001593 UP000008237 UP000242188 UP000002320 UP000091820 UP000092444 UP000092443 UP000092445 UP000261560 UP000085678 UP000009192 UP000092460 UP000008744 UP000078200 UP000001819 UP000186698 UP000015103 UP000261520 UP000053605 UP000261420 UP000016665 UP000265020 UP000190648 UP000230750
Interpro
IPR013848
Methylthiotransferase_N
+ More
IPR002792 TRAM_dom
IPR020612 Methylthiotransferase_CS
IPR038135 Methylthiotransferase_N_sf
IPR005839 Methylthiotransferase
IPR023404 rSAM_horseshoe
IPR007197 rSAM
IPR006466 MiaB-like_B
IPR006638 Elp3/MiaB/NifB
IPR036237 Xyl_isomerase-like_sf
IPR001998 Xylose_isomerase
IPR032198 E2F_CC-MB
IPR037241 E2F-DP_heterodim
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR003316 E2F_WHTH_DNA-bd_dom
IPR002792 TRAM_dom
IPR020612 Methylthiotransferase_CS
IPR038135 Methylthiotransferase_N_sf
IPR005839 Methylthiotransferase
IPR023404 rSAM_horseshoe
IPR007197 rSAM
IPR006466 MiaB-like_B
IPR006638 Elp3/MiaB/NifB
IPR036237 Xyl_isomerase-like_sf
IPR001998 Xylose_isomerase
IPR032198 E2F_CC-MB
IPR037241 E2F-DP_heterodim
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR003316 E2F_WHTH_DNA-bd_dom
Gene 3D
CDD
ProteinModelPortal
H9JR09
A0A194Q181
A0A2A4JIL5
A0A2H1VNN1
S4PM93
F1C4C0
+ More
A0A2W1BYK8 A0A212FDU8 D6W6D0 A0A151J017 A0A026WQS6 E9ICL2 A0A195FNM0 A0A2J7PJS5 A0A195CVR9 E1ZVZ2 A0A0M8ZXM5 A0A2A3ESV7 A0A0J7L5J6 A0A154P5T8 F4WMT0 A0A1Y1M1F3 A0A0L7QPS3 A0A195BJ44 A0A158NQ43 U4U8D7 E9FR10 B4NQ57 A0A0P5N242 A0A3B0JMC1 A0A0P5VGW0 A0A0P6HHG8 A0A0P5AZR9 A0A3B0JUD9 A0A0P6F6N9 A0A0P4YGR0 B3MG65 A0A0P5VCT9 A0A0P5U398 A0A1B6CSR2 A0A1W4VDB4 A0A1B0CY58 B4LPW9 B4J918 A0A1B0DK13 B3NNR9 A0A0P4WFE8 Q7K4W1 A0A1B6KGQ2 A0A232EI88 B4QAT0 B4HML1 A0A1I8NAH0 A0A1B0DFB9 A0A0P5PKE8 Q16Q27 A0A1I8NZW7 A0A0M4EU02 A7S7F5 A0A0K8TM36 E2BHJ6 A0A1Q3F360 A0A1B6EL38 A0A1B6G2W2 A0A210PNT2 B0W0A5 A0A1A9WNI8 W8BFT4 A0A1B0G4Q9 A0A023F4W3 A0A1A9Y303 A0A1A9ZKW6 V9KMV6 A0A069DUI9 A0A3B3DID5 A0A1S3IFX3 B4KQZ9 A0A1B0C2H0 B4GAQ6 A0A3B3DIE0 A0A3B3C2A1 A0A1A9VKQ5 Q291H5 A0A1L8FSF7 A0A224XM19 A0A0S7FQS5 Q6NS26 T1ICA5 A0A3B4BDX2 A0A091WK03 A0A3B4UQR5 A0A3B4URE2 U3KGF0 A0A3B4UT16 A0A0A1WDX7 A0A3Q2CNL6 A0A1V4JGI9 R0LT49 A0A1A8UAR2 A0A2G8JTL9
A0A2W1BYK8 A0A212FDU8 D6W6D0 A0A151J017 A0A026WQS6 E9ICL2 A0A195FNM0 A0A2J7PJS5 A0A195CVR9 E1ZVZ2 A0A0M8ZXM5 A0A2A3ESV7 A0A0J7L5J6 A0A154P5T8 F4WMT0 A0A1Y1M1F3 A0A0L7QPS3 A0A195BJ44 A0A158NQ43 U4U8D7 E9FR10 B4NQ57 A0A0P5N242 A0A3B0JMC1 A0A0P5VGW0 A0A0P6HHG8 A0A0P5AZR9 A0A3B0JUD9 A0A0P6F6N9 A0A0P4YGR0 B3MG65 A0A0P5VCT9 A0A0P5U398 A0A1B6CSR2 A0A1W4VDB4 A0A1B0CY58 B4LPW9 B4J918 A0A1B0DK13 B3NNR9 A0A0P4WFE8 Q7K4W1 A0A1B6KGQ2 A0A232EI88 B4QAT0 B4HML1 A0A1I8NAH0 A0A1B0DFB9 A0A0P5PKE8 Q16Q27 A0A1I8NZW7 A0A0M4EU02 A7S7F5 A0A0K8TM36 E2BHJ6 A0A1Q3F360 A0A1B6EL38 A0A1B6G2W2 A0A210PNT2 B0W0A5 A0A1A9WNI8 W8BFT4 A0A1B0G4Q9 A0A023F4W3 A0A1A9Y303 A0A1A9ZKW6 V9KMV6 A0A069DUI9 A0A3B3DID5 A0A1S3IFX3 B4KQZ9 A0A1B0C2H0 B4GAQ6 A0A3B3DIE0 A0A3B3C2A1 A0A1A9VKQ5 Q291H5 A0A1L8FSF7 A0A224XM19 A0A0S7FQS5 Q6NS26 T1ICA5 A0A3B4BDX2 A0A091WK03 A0A3B4UQR5 A0A3B4URE2 U3KGF0 A0A3B4UT16 A0A0A1WDX7 A0A3Q2CNL6 A0A1V4JGI9 R0LT49 A0A1A8UAR2 A0A2G8JTL9
PDB
4JC0
E-value=1.35291e-25,
Score=290
Ontologies
GO
Topology
Subcellular location
Membrane
Endoplasmic reticulum membrane
Nucleus
Endoplasmic reticulum membrane
Nucleus
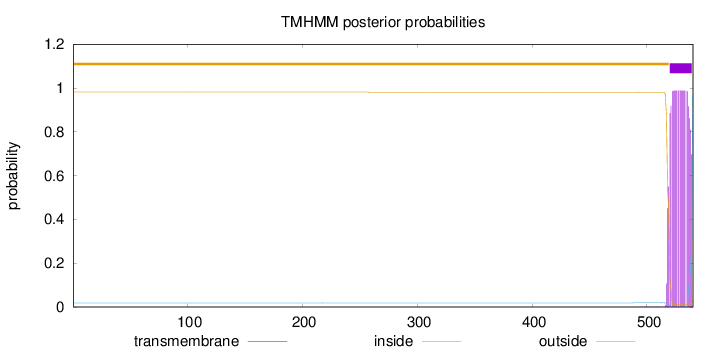
Length:
540
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.0444
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01860
outside
1 - 519
TMhelix
520 - 539
inside
540 - 540
Population Genetic Test Statistics
Pi
179.104753
Theta
165.620972
Tajima's D
0.596461
CLR
0.274937
CSRT
0.544222788860557
Interpretation
Uncertain
