Pre Gene Modal
BGIBMGA011932
Annotation
PREDICTED:_uridine-cytidine_kinase-like_1_isoform_X2_[Bombyx_mori]
Full name
Uridine kinase
Location in the cell
Cytoplasmic Reliability : 1.409 Mitochondrial Reliability : 1.702
Sequence
CDS
ATGGCTGGGAAATTGAACAAATTCGAAACGCCCAGCTCTGCCAGCTCTGATAGCGACACTGAATTAGCGTTAGAGTTAAGCGAACAAGTATTCCTTGACGATGAAGACTACTCGAGCAGAGCTCCCGCCTCGCCCACAACTGTACCGAACGTGCGACCATACAGACCTCCGTCCACCGGATCGAATATACTGAAGTCTCCGCGAGCGAGGCGCGTCCGCACAACGTCAATGTCACAGTCAAACAAACGCACAGCGGCCGAGAGCATTCTTCATGCTGACAAAAGGACTATATACACCGCCGGCAGACCCCCCTGGTACAACTGTACAGGGGGGCAGGAGGTGGAACCCTTTCTTATAGGTATATGCGGCGCGAGTGCGTCCGGCAAGACGACAGTGGCTGCCAAAATCATAGAATCTCTCAACATACCCTGGGTCACTATCGTCAGCATGGATTCATTTTACAAAGTACTTAACGAGAAACAGCACCACCTGGCGGCCCTCAACGAATATAACTTCGACCACCCTGACGCCTTCGACATGGAGTTGCTGGTTAGCGTGCTGCAGCGGCTTCGCGAGGGAAAGAAAGTGGAAGTTCCGATATACAACTACGTCACGCACGCGAGGGAGAATAGAACTAAAACAATGTACGGCGCGAACGTGATAATATTTGAAGGCATTCTGGCCTTCTACAACGCGGAGGTGCTGAAGCTTCTGGACATGAAGGTGTTCGTGGACACGGACGCCGACATACGGCTCGCCAGGAGACTGCGCAGGGACATCGTGCAGCGCGGGCGCGAGCTGGAGGGCGTGCTGAAGCAGTACATGACGTACGTGAAGCCCGCCTACCAGAACTACATCGCGCCGTGCATGGCGCACGCCGACATCATCGTGCCGCGCGGCGGGGAGAACAAGGTCGCCATCCACCTCATCGTGCAGCACGTGCACAAGCAGCTGCAGCTGCGCGGGTTCAAGGTGCGCGAGAAGCTGGCGGTGGCGCACGTGGGCCAGCCCGTGCCCGACTCGCTCTACGTGCTCGAGAACACGCCGCAGGTGCAGGGGCTCCACACGTTCATACGCAACAAGGACACGCCGCGGGACGAGTTCATATTCTACTCTAAGCGCCTGATGCGCCTCGTCATAGAGTTCGCGCTGTCGCTGCAGCCCTACGCGCCGCGCACCGTGGACACGCCGCAGGGGATCGCCTACCACGGTCGGAAGTGCGAGGTGGAGAAGATATGTGGCGTGTCCATCCTGCGCGCCGGTGAGACCATGGAGCAGGCCGTGTGCGACGTGTGCAAGGACATCCGCATCGGGAAGATACTCATCCAGACCAACCAGCTCACGGACGAGCCCGAGCTCTACTACCTGCGGCTGCCCAAGGACATCAAGGACTACCGCGTGATCCTGATGGACGCGACGGTGGCGACGGGCGCGGCCGCCATCATGGCCATCCGCGTGCTGCTGGACCACGACGTGCCGGAGCACTGCATCAGCCTCGTGTCGCTGCTCATGGCGGACGCGGGCGCGCACTCCATCGCCTACGCCTTCCCGCAGGTAAAGATCGTAACGTCGGCCCTGGATCCCGAGATCAACGAGAAGTTCTACGTGCTGCCCGGGATAGGCAACTTCGGCGACCGGTATTTCGGCACCGAGCCCGCCGACGACGAGTGA
Protein
MAGKLNKFETPSSASSDSDTELALELSEQVFLDDEDYSSRAPASPTTVPNVRPYRPPSTGSNILKSPRARRVRTTSMSQSNKRTAAESILHADKRTIYTAGRPPWYNCTGGQEVEPFLIGICGASASGKTTVAAKIIESLNIPWVTIVSMDSFYKVLNEKQHHLAALNEYNFDHPDAFDMELLVSVLQRLREGKKVEVPIYNYVTHARENRTKTMYGANVIIFEGILAFYNAEVLKLLDMKVFVDTDADIRLARRLRRDIVQRGRELEGVLKQYMTYVKPAYQNYIAPCMAHADIIVPRGGENKVAIHLIVQHVHKQLQLRGFKVREKLAVAHVGQPVPDSLYVLENTPQVQGLHTFIRNKDTPRDEFIFYSKRLMRLVIEFALSLQPYAPRTVDTPQGIAYHGRKCEVEKICGVSILRAGETMEQAVCDVCKDIRIGKILIQTNQLTDEPELYYLRLPKDIKDYRVILMDATVATGAAAIMAIRVLLDHDVPEHCISLVSLLMADAGAHSIAYAFPQVKIVTSALDPEINEKFYVLPGIGNFGDRYFGTEPADDE
Summary
Catalytic Activity
ATP + cytidine = ADP + CMP + H(+)
ATP + uridine = ADP + H(+) + UMP
ATP + uridine = ADP + H(+) + UMP
Similarity
Belongs to the uridine kinase family.
Feature
chain Uridine kinase
Uniprot
A0A2A4JHW5
A0A3S2LIH0
A0A2A4JIQ3
A0A212FDW7
A0A2Z5UIM3
A0A194PWM0
+ More
A0A2W1BYF4 H9JQX6 A0A2J7PJS0 A0A0C9QP02 A0A026WLZ0 E9I9V6 A0A195CJP8 E2AT07 A0A195E5M2 F4W458 A0A195B479 A0A158NF19 A0A151X266 A0A195ETA6 A0A067R085 A0A232F2R1 A0A0L7LBM3 A0A0M9A684 D6W6C4 A0A2J7PJS1 U5ESP1 E2B6P7 A0A1B6DMB5 A0A154PCA0 A0A1B6E793 A0A0K8TSP4 A0A2S2RAF4 B0X066 A0A1S4FST9 A0A1B6HKH6 A0A0A9YDW2 A0A2H8TJC6 Q16QI9 A0A023EUT9 A0A023F5K1 A0A069DVN4 A0A2S2P4P2 X1X1Y2 A0A310SUW7 A0A0P4VUD7 A0A224XMR6 A0A1B6L5I5 W8BGJ9 A0A1Q3FDI4 A0A1Y1L7I3 A0A034W8B3 A0A1L8DJ67 A0A034W9U3 W8C3B7 A0A034W4M1 W8BTX0 W8CA20 A0A1W4XJC2 B3MG64 B4MQJ0 A0A1B6EV68 A0A0P8XP30 A0A182QYB9 K7J2T4 A0A1W4XUD2 A0A087ZVQ9 A0A1W4VDA9 A0A084WJI7 A0A1W4VBZ4 A0A1W4V040 B4LPX0 B4KR00 A0A182RKV3 A0A3B0JH38 B4J917 A0A182P8R9 A0A182IQC1 A0A182XEW0 Q5TSQ7 A0A182HNY5 A0A182NQ55 A0A182YC46 B3NNS0 A0A0L7QSW4 A0A182K8H8 A0A0Q5VXT3 A0A182WK29 B4P807 B5E031 B4GAQ5 Q8MQK4 A0A0R1DT41 A0A2M4BJI2 A0A0R3NLZ1 A0A0R3NSZ0 A1ZAS3 A0A2M4AFR4 A0A2M3Z7Y4 A0A182FCY4
A0A2W1BYF4 H9JQX6 A0A2J7PJS0 A0A0C9QP02 A0A026WLZ0 E9I9V6 A0A195CJP8 E2AT07 A0A195E5M2 F4W458 A0A195B479 A0A158NF19 A0A151X266 A0A195ETA6 A0A067R085 A0A232F2R1 A0A0L7LBM3 A0A0M9A684 D6W6C4 A0A2J7PJS1 U5ESP1 E2B6P7 A0A1B6DMB5 A0A154PCA0 A0A1B6E793 A0A0K8TSP4 A0A2S2RAF4 B0X066 A0A1S4FST9 A0A1B6HKH6 A0A0A9YDW2 A0A2H8TJC6 Q16QI9 A0A023EUT9 A0A023F5K1 A0A069DVN4 A0A2S2P4P2 X1X1Y2 A0A310SUW7 A0A0P4VUD7 A0A224XMR6 A0A1B6L5I5 W8BGJ9 A0A1Q3FDI4 A0A1Y1L7I3 A0A034W8B3 A0A1L8DJ67 A0A034W9U3 W8C3B7 A0A034W4M1 W8BTX0 W8CA20 A0A1W4XJC2 B3MG64 B4MQJ0 A0A1B6EV68 A0A0P8XP30 A0A182QYB9 K7J2T4 A0A1W4XUD2 A0A087ZVQ9 A0A1W4VDA9 A0A084WJI7 A0A1W4VBZ4 A0A1W4V040 B4LPX0 B4KR00 A0A182RKV3 A0A3B0JH38 B4J917 A0A182P8R9 A0A182IQC1 A0A182XEW0 Q5TSQ7 A0A182HNY5 A0A182NQ55 A0A182YC46 B3NNS0 A0A0L7QSW4 A0A182K8H8 A0A0Q5VXT3 A0A182WK29 B4P807 B5E031 B4GAQ5 Q8MQK4 A0A0R1DT41 A0A2M4BJI2 A0A0R3NLZ1 A0A0R3NSZ0 A1ZAS3 A0A2M4AFR4 A0A2M3Z7Y4 A0A182FCY4
EC Number
2.7.1.48
Pubmed
22118469
26354079
28756777
19121390
24508170
30249741
+ More
21282665 20798317 21719571 21347285 24845553 28648823 26227816 18362917 19820115 26369729 25401762 26823975 17510324 24945155 26483478 25474469 26334808 27129103 24495485 28004739 25348373 17994087 20075255 24438588 12364791 14747013 17210077 25244985 18057021 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
21282665 20798317 21719571 21347285 24845553 28648823 26227816 18362917 19820115 26369729 25401762 26823975 17510324 24945155 26483478 25474469 26334808 27129103 24495485 28004739 25348373 17994087 20075255 24438588 12364791 14747013 17210077 25244985 18057021 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
NWSH01001334
PCG71647.1
RSAL01000104
RVE47368.1
PCG71646.1
AGBW02008998
+ More
OWR51898.1 AP017476 BBB06712.1 KQ459590 KPI97154.1 KZ149897 PZC78685.1 BABH01028575 NEVH01024946 PNF16587.1 GBYB01005279 JAG75046.1 KK107153 QOIP01000012 EZA56983.1 RLU15811.1 GL761846 EFZ22673.1 KQ977642 KYN00955.1 GL442439 EFN63442.1 KQ979609 KYN20149.1 GL887491 EGI71054.1 KQ976625 KYM79014.1 ADTU01001499 KQ982585 KYQ54284.1 KQ981979 KYN31485.1 KK852833 KDR15301.1 NNAY01001153 OXU24975.1 JTDY01001892 KOB72616.1 KQ435727 KOX78077.1 KQ971319 EFA11381.2 PNF16586.1 GANO01003152 JAB56719.1 GL446000 EFN88631.1 GEDC01010442 JAS26856.1 KQ434870 KZC09526.1 GEDC01003574 JAS33724.1 GDAI01000638 JAI16965.1 GGMS01017447 MBY86650.1 DS232229 EDS37941.1 GECU01032523 JAS75183.1 GBHO01014311 GBHO01014309 GBRD01008538 GBRD01008537 GDHC01009665 JAG29293.1 JAG29295.1 JAG57283.1 JAQ08964.1 GFXV01002411 MBW14216.1 CH477747 EAT36651.1 JXUM01074255 JXUM01074256 GAPW01000857 KQ562819 JAC12741.1 KXJ75061.1 GBBI01002272 JAC16440.1 GBGD01001033 JAC87856.1 GGMR01011820 MBY24439.1 ABLF02031447 ABLF02031448 KQ760208 OAD61380.1 GDKW01000949 JAI55646.1 GFTR01007137 JAW09289.1 GEBQ01020998 JAT18979.1 GAMC01006155 JAC00401.1 GFDL01009432 JAV25613.1 GEZM01066410 GEZM01066409 JAV67905.1 GAKP01008405 GAKP01008403 JAC50547.1 GFDF01007576 JAV06508.1 GAKP01008404 GAKP01008402 JAC50548.1 GAMC01006154 GAMC01006150 JAC00402.1 GAKP01008401 JAC50551.1 GAMC01006152 JAC00404.1 GAMC01006151 JAC00405.1 CH902619 EDV36759.1 CH963849 EDW74379.1 GECZ01027999 JAS41770.1 KPU76399.1 AXCN02001020 ATLV01024035 KE525348 KFB50381.1 CH940648 EDW61310.2 CH933808 EDW10356.2 OUUW01000001 SPP74690.1 CH916367 EDW01366.1 AAAB01008900 EAL40491.3 APCN01002812 CH954179 EDV55627.1 KQ414755 KOC61743.1 KQS62350.1 CM000158 EDW92162.1 CM000071 EDY68915.1 CH479181 EDW32007.1 AE013599 AY129436 AAF57871.3 AAM76178.1 KRK00348.1 GGFJ01003807 MBW52948.1 KRT01874.1 KRT01875.1 BT125900 AAF57873.2 ADV19041.1 GGFK01006303 MBW39624.1 GGFM01003849 MBW24600.1
OWR51898.1 AP017476 BBB06712.1 KQ459590 KPI97154.1 KZ149897 PZC78685.1 BABH01028575 NEVH01024946 PNF16587.1 GBYB01005279 JAG75046.1 KK107153 QOIP01000012 EZA56983.1 RLU15811.1 GL761846 EFZ22673.1 KQ977642 KYN00955.1 GL442439 EFN63442.1 KQ979609 KYN20149.1 GL887491 EGI71054.1 KQ976625 KYM79014.1 ADTU01001499 KQ982585 KYQ54284.1 KQ981979 KYN31485.1 KK852833 KDR15301.1 NNAY01001153 OXU24975.1 JTDY01001892 KOB72616.1 KQ435727 KOX78077.1 KQ971319 EFA11381.2 PNF16586.1 GANO01003152 JAB56719.1 GL446000 EFN88631.1 GEDC01010442 JAS26856.1 KQ434870 KZC09526.1 GEDC01003574 JAS33724.1 GDAI01000638 JAI16965.1 GGMS01017447 MBY86650.1 DS232229 EDS37941.1 GECU01032523 JAS75183.1 GBHO01014311 GBHO01014309 GBRD01008538 GBRD01008537 GDHC01009665 JAG29293.1 JAG29295.1 JAG57283.1 JAQ08964.1 GFXV01002411 MBW14216.1 CH477747 EAT36651.1 JXUM01074255 JXUM01074256 GAPW01000857 KQ562819 JAC12741.1 KXJ75061.1 GBBI01002272 JAC16440.1 GBGD01001033 JAC87856.1 GGMR01011820 MBY24439.1 ABLF02031447 ABLF02031448 KQ760208 OAD61380.1 GDKW01000949 JAI55646.1 GFTR01007137 JAW09289.1 GEBQ01020998 JAT18979.1 GAMC01006155 JAC00401.1 GFDL01009432 JAV25613.1 GEZM01066410 GEZM01066409 JAV67905.1 GAKP01008405 GAKP01008403 JAC50547.1 GFDF01007576 JAV06508.1 GAKP01008404 GAKP01008402 JAC50548.1 GAMC01006154 GAMC01006150 JAC00402.1 GAKP01008401 JAC50551.1 GAMC01006152 JAC00404.1 GAMC01006151 JAC00405.1 CH902619 EDV36759.1 CH963849 EDW74379.1 GECZ01027999 JAS41770.1 KPU76399.1 AXCN02001020 ATLV01024035 KE525348 KFB50381.1 CH940648 EDW61310.2 CH933808 EDW10356.2 OUUW01000001 SPP74690.1 CH916367 EDW01366.1 AAAB01008900 EAL40491.3 APCN01002812 CH954179 EDV55627.1 KQ414755 KOC61743.1 KQS62350.1 CM000158 EDW92162.1 CM000071 EDY68915.1 CH479181 EDW32007.1 AE013599 AY129436 AAF57871.3 AAM76178.1 KRK00348.1 GGFJ01003807 MBW52948.1 KRT01874.1 KRT01875.1 BT125900 AAF57873.2 ADV19041.1 GGFK01006303 MBW39624.1 GGFM01003849 MBW24600.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000005204
UP000235965
+ More
UP000053097 UP000279307 UP000078542 UP000000311 UP000078492 UP000007755 UP000078540 UP000005205 UP000075809 UP000078541 UP000027135 UP000215335 UP000037510 UP000053105 UP000007266 UP000008237 UP000076502 UP000002320 UP000008820 UP000069940 UP000249989 UP000007819 UP000192223 UP000007801 UP000007798 UP000075886 UP000002358 UP000005203 UP000192221 UP000030765 UP000008792 UP000009192 UP000075900 UP000268350 UP000001070 UP000075885 UP000075880 UP000076407 UP000007062 UP000075840 UP000075884 UP000076408 UP000008711 UP000053825 UP000075881 UP000075920 UP000002282 UP000001819 UP000008744 UP000000803 UP000069272
UP000053097 UP000279307 UP000078542 UP000000311 UP000078492 UP000007755 UP000078540 UP000005205 UP000075809 UP000078541 UP000027135 UP000215335 UP000037510 UP000053105 UP000007266 UP000008237 UP000076502 UP000002320 UP000008820 UP000069940 UP000249989 UP000007819 UP000192223 UP000007801 UP000007798 UP000075886 UP000002358 UP000005203 UP000192221 UP000030765 UP000008792 UP000009192 UP000075900 UP000268350 UP000001070 UP000075885 UP000075880 UP000076407 UP000007062 UP000075840 UP000075884 UP000076408 UP000008711 UP000053825 UP000075881 UP000075920 UP000002282 UP000001819 UP000008744 UP000000803 UP000069272
Pfam
PF00485 PRK
Interpro
ProteinModelPortal
A0A2A4JHW5
A0A3S2LIH0
A0A2A4JIQ3
A0A212FDW7
A0A2Z5UIM3
A0A194PWM0
+ More
A0A2W1BYF4 H9JQX6 A0A2J7PJS0 A0A0C9QP02 A0A026WLZ0 E9I9V6 A0A195CJP8 E2AT07 A0A195E5M2 F4W458 A0A195B479 A0A158NF19 A0A151X266 A0A195ETA6 A0A067R085 A0A232F2R1 A0A0L7LBM3 A0A0M9A684 D6W6C4 A0A2J7PJS1 U5ESP1 E2B6P7 A0A1B6DMB5 A0A154PCA0 A0A1B6E793 A0A0K8TSP4 A0A2S2RAF4 B0X066 A0A1S4FST9 A0A1B6HKH6 A0A0A9YDW2 A0A2H8TJC6 Q16QI9 A0A023EUT9 A0A023F5K1 A0A069DVN4 A0A2S2P4P2 X1X1Y2 A0A310SUW7 A0A0P4VUD7 A0A224XMR6 A0A1B6L5I5 W8BGJ9 A0A1Q3FDI4 A0A1Y1L7I3 A0A034W8B3 A0A1L8DJ67 A0A034W9U3 W8C3B7 A0A034W4M1 W8BTX0 W8CA20 A0A1W4XJC2 B3MG64 B4MQJ0 A0A1B6EV68 A0A0P8XP30 A0A182QYB9 K7J2T4 A0A1W4XUD2 A0A087ZVQ9 A0A1W4VDA9 A0A084WJI7 A0A1W4VBZ4 A0A1W4V040 B4LPX0 B4KR00 A0A182RKV3 A0A3B0JH38 B4J917 A0A182P8R9 A0A182IQC1 A0A182XEW0 Q5TSQ7 A0A182HNY5 A0A182NQ55 A0A182YC46 B3NNS0 A0A0L7QSW4 A0A182K8H8 A0A0Q5VXT3 A0A182WK29 B4P807 B5E031 B4GAQ5 Q8MQK4 A0A0R1DT41 A0A2M4BJI2 A0A0R3NLZ1 A0A0R3NSZ0 A1ZAS3 A0A2M4AFR4 A0A2M3Z7Y4 A0A182FCY4
A0A2W1BYF4 H9JQX6 A0A2J7PJS0 A0A0C9QP02 A0A026WLZ0 E9I9V6 A0A195CJP8 E2AT07 A0A195E5M2 F4W458 A0A195B479 A0A158NF19 A0A151X266 A0A195ETA6 A0A067R085 A0A232F2R1 A0A0L7LBM3 A0A0M9A684 D6W6C4 A0A2J7PJS1 U5ESP1 E2B6P7 A0A1B6DMB5 A0A154PCA0 A0A1B6E793 A0A0K8TSP4 A0A2S2RAF4 B0X066 A0A1S4FST9 A0A1B6HKH6 A0A0A9YDW2 A0A2H8TJC6 Q16QI9 A0A023EUT9 A0A023F5K1 A0A069DVN4 A0A2S2P4P2 X1X1Y2 A0A310SUW7 A0A0P4VUD7 A0A224XMR6 A0A1B6L5I5 W8BGJ9 A0A1Q3FDI4 A0A1Y1L7I3 A0A034W8B3 A0A1L8DJ67 A0A034W9U3 W8C3B7 A0A034W4M1 W8BTX0 W8CA20 A0A1W4XJC2 B3MG64 B4MQJ0 A0A1B6EV68 A0A0P8XP30 A0A182QYB9 K7J2T4 A0A1W4XUD2 A0A087ZVQ9 A0A1W4VDA9 A0A084WJI7 A0A1W4VBZ4 A0A1W4V040 B4LPX0 B4KR00 A0A182RKV3 A0A3B0JH38 B4J917 A0A182P8R9 A0A182IQC1 A0A182XEW0 Q5TSQ7 A0A182HNY5 A0A182NQ55 A0A182YC46 B3NNS0 A0A0L7QSW4 A0A182K8H8 A0A0Q5VXT3 A0A182WK29 B4P807 B5E031 B4GAQ5 Q8MQK4 A0A0R1DT41 A0A2M4BJI2 A0A0R3NLZ1 A0A0R3NSZ0 A1ZAS3 A0A2M4AFR4 A0A2M3Z7Y4 A0A182FCY4
PDB
1UJ2
E-value=2.85334e-53,
Score=529
Ontologies
PATHWAY
GO
PANTHER
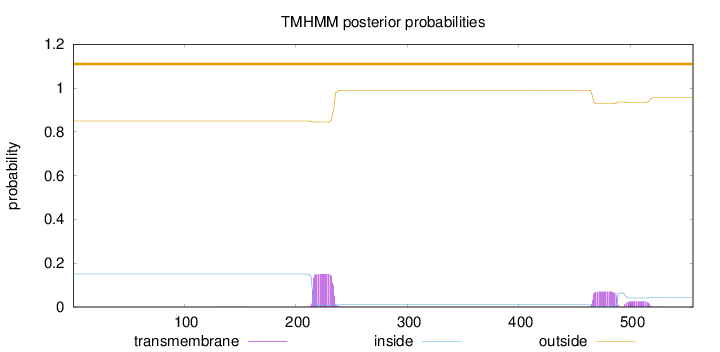
Topology
Length:
556
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.03063
Exp number, first 60 AAs:
0
Total prob of N-in:
0.15054
outside
1 - 556
Population Genetic Test Statistics
Pi
225.027914
Theta
184.778607
Tajima's D
1.048363
CLR
0.488905
CSRT
0.672016399180041
Interpretation
Uncertain
