Gene
KWMTBOMO06396
Pre Gene Modal
BGIBMGA011931
Annotation
PREDICTED:_netrin-B_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.841 Nuclear Reliability : 1.948
Sequence
CDS
ATGGCGGAATGCTCGTGCAATGGCCACGCTCGGCGGTGCCGGTTCAACATGGAGCTTTACAAGCTGTCGGGCCGCGACAGTGGCGGCGTATGCCTTAAATGCCGGCATTACACGGCCGGCAGGCATTGCCATTACTGCCGGGAAGGATACTACAGGGATCCTACCAAGCCCATCACTCACAAGAAGGCTTGCAAACCCTGCGACTGTCACCCTGTCGGAGCCAGTGGGAAGACCTGCAATCAGACGAGCGGTCAGTGTCCTTGCAAGGATGGCGTGACAGGGACCACGTGCAATAGATGCGCCAAAGGATACCAACAGTCCAGGAGTCATATTGCGCCTTGCATACGTATACCTCGTGTGGTGACTGCTGTACAGGCTATGGAAGCCGAAGACAACGAGCCGGGCCGCGCAGCGGACCAGTGCGGTCGGTGTCGCGTCGGCGCCCGCACGCTAAACCTGAACAAGTTCTGCAGCCGAGACTACGTCATAATGGGCAAGGTGGTAGGTCGCGAGGCGAGCGCGGCTGGCGGCGCCTGGTCCCGTCTGGCGCTCAACGTGCAGGCGGTGTACAAGCGCGCTCCCCGCAGCCGGCTGCGACGGGGCGCCGCCGCGCTCTTCGTGCGCGCCCGGGACCTCGCCTGCAAGTGTCCTAAACTAAAGATCAACAAATCATACCTAATCCTAGGAGTGGAAAAAGAAGGTCTTCAAGCTGGCTTACCGGGCTTAGTGGTGGGAGAGAGAACCCTTCTCCTTGAGTGGCGCGACGACTGGCACCGACGCATCCGTCGTCTTCAGCGTCGCGCGATCAACTGCCACTAG
Protein
MAECSCNGHARRCRFNMELYKLSGRDSGGVCLKCRHYTAGRHCHYCREGYYRDPTKPITHKKACKPCDCHPVGASGKTCNQTSGQCPCKDGVTGTTCNRCAKGYQQSRSHIAPCIRIPRVVTAVQAMEAEDNEPGRAADQCGRCRVGARTLNLNKFCSRDYVIMGKVVGREASAAGGAWSRLALNVQAVYKRAPRSRLRRGAAALFVRARDLACKCPKLKINKSYLILGVEKEGLQAGLPGLVVGERTLLLEWRDDWHRRIRRLQRRAINCH
Summary
Uniprot
H9JQX5
A0A2W1BZJ7
A0A194PV02
A0A0N1PHN9
A0A3S2TJ02
A0A212FDR4
+ More
A0A1B6CV77 A0A1B6J0X0 A0A0L7LU79 A0A1B6L4V9 A0A158NJF8 A0A067RDY5 A0A0C9PLN5 A0A1Y1M261 A0A154PNU8 A0A1B6EZB2 E2BUA0 A0A1Y1M907 D7EHQ2 A0A0A9Z2X5 E0VYV4 A0A1Y1M443 A0A0P5JJJ3 A0A2S2QES5 A0A146MHX1 A0A0P4XQH6 A0A0P5RCR6 A0A0P6HWB0 A0A0N7ZFE1 A0A0M9ABD1 A0A2S2P501 A0A2H8TME8 A0A182MGN3 E9GZG2 A0A226ECW7 A0A182PN48 A0A1B0C9C2 A0A182IAE8 A0A1D2MQP3 A0A182JHX5 A0A182WTN4 A0A182V2Z4 T1K3U5 T1DI33 A0A3L8DF10 A0A182QSJ5 A0A182WQJ7 A0A182S4K3 A0A2P1DV55 A0A182NZ13 K7J7Q0 Q0IGC3 A0A1I8N3U9 A0A0P5DCN6 L5K039 G7PTM1 U6D9U2 A0A1Q3FV05 A0A1Q3FV72 W5UHM1 Q0IGC8 V9KAW1 W5KQW5 V9K9V4 A0A146WJ74 A0A1L8EV13 A0A3Q1INQ5 G1KUB3 A0A2U9BQD3 O42140 A0A0B5JPZ7 A0A1W5A9T8 F7CBX5 A0A3B4WFQ0 A0A3B4VCM8 A0A3B3XTX2 A0A3Q2YN80 A0A1S3PS44 A0A3B5B6R7 A0A3B3W2Z4 A0A3Q1B3Y3 F7G4D6 A0A3P8XKC4 G3Q6U6 A0A2R9AA25 K9ITB3 B4JJN0 B3N1K3 Q95SP5 O57339 M3ZWP7 A0A0M5JDR0 L8HV67 A0A2K5ZVT1 A0A2K6SXG2 B4GY45 A0A3B1KE85 A0A1S3L5W0 A0A3P8RME6
A0A1B6CV77 A0A1B6J0X0 A0A0L7LU79 A0A1B6L4V9 A0A158NJF8 A0A067RDY5 A0A0C9PLN5 A0A1Y1M261 A0A154PNU8 A0A1B6EZB2 E2BUA0 A0A1Y1M907 D7EHQ2 A0A0A9Z2X5 E0VYV4 A0A1Y1M443 A0A0P5JJJ3 A0A2S2QES5 A0A146MHX1 A0A0P4XQH6 A0A0P5RCR6 A0A0P6HWB0 A0A0N7ZFE1 A0A0M9ABD1 A0A2S2P501 A0A2H8TME8 A0A182MGN3 E9GZG2 A0A226ECW7 A0A182PN48 A0A1B0C9C2 A0A182IAE8 A0A1D2MQP3 A0A182JHX5 A0A182WTN4 A0A182V2Z4 T1K3U5 T1DI33 A0A3L8DF10 A0A182QSJ5 A0A182WQJ7 A0A182S4K3 A0A2P1DV55 A0A182NZ13 K7J7Q0 Q0IGC3 A0A1I8N3U9 A0A0P5DCN6 L5K039 G7PTM1 U6D9U2 A0A1Q3FV05 A0A1Q3FV72 W5UHM1 Q0IGC8 V9KAW1 W5KQW5 V9K9V4 A0A146WJ74 A0A1L8EV13 A0A3Q1INQ5 G1KUB3 A0A2U9BQD3 O42140 A0A0B5JPZ7 A0A1W5A9T8 F7CBX5 A0A3B4WFQ0 A0A3B4VCM8 A0A3B3XTX2 A0A3Q2YN80 A0A1S3PS44 A0A3B5B6R7 A0A3B3W2Z4 A0A3Q1B3Y3 F7G4D6 A0A3P8XKC4 G3Q6U6 A0A2R9AA25 K9ITB3 B4JJN0 B3N1K3 Q95SP5 O57339 M3ZWP7 A0A0M5JDR0 L8HV67 A0A2K5ZVT1 A0A2K6SXG2 B4GY45 A0A3B1KE85 A0A1S3L5W0 A0A3P8RME6
Pubmed
19121390
28756777
26354079
22118469
26227816
21347285
+ More
24845553 28004739 20798317 18362917 19820115 25401762 20566863 26823975 21292972 27289101 30249741 29236686 20075255 17510324 25315136 23258410 22002653 23127152 24402279 25329095 27762356 9268507 23594743 26190107 20431018 17495919 25069045 22722832 17994087 9427245 23542700 22751099
24845553 28004739 20798317 18362917 19820115 25401762 20566863 26823975 21292972 27289101 30249741 29236686 20075255 17510324 25315136 23258410 22002653 23127152 24402279 25329095 27762356 9268507 23594743 26190107 20431018 17495919 25069045 22722832 17994087 9427245 23542700 22751099
EMBL
BABH01028583
BABH01028584
KZ149897
PZC78687.1
KQ459590
KPI97152.1
+ More
KQ460210 KPJ16592.1 RSAL01000104 RVE47366.1 AGBW02008998 OWR51896.1 GEDC01020017 JAS17281.1 GECU01019739 GECU01014871 JAS87967.1 JAS92835.1 JTDY01000076 KOB79002.1 GEBQ01021277 JAT18700.1 ADTU01001922 ADTU01001923 ADTU01001924 ADTU01001925 ADTU01001926 ADTU01001927 ADTU01001928 ADTU01001929 ADTU01001930 ADTU01001931 KK852567 KDR21223.1 GBYB01001992 GBYB01001993 JAG71759.1 JAG71760.1 GEZM01042509 JAV79809.1 KQ435005 KZC13407.1 GECZ01026511 JAS43258.1 GL450619 EFN80728.1 GEZM01042511 JAV79807.1 KQ973114 EFA12173.2 GBHO01005368 JAG38236.1 DS235848 EEB18560.1 GEZM01042510 JAV79808.1 GDIQ01204138 JAK47587.1 GGMS01007042 MBY76245.1 GDHC01000349 JAQ18280.1 GDIP01240540 JAI82861.1 GDIQ01122073 JAL29653.1 GDIQ01021178 JAN73559.1 GDIP01250809 JAI72592.1 KQ435692 KOX81075.1 GGMR01011803 MBY24422.1 GFXV01003306 MBW15111.1 AXCM01009261 GL732577 EFX75164.1 LNIX01000004 OXA55423.1 AJWK01002192 AJWK01002193 AJWK01002194 APCN01001217 LJIJ01000678 ODM95397.1 CAEY01001560 GAMD01002071 JAA99519.1 QOIP01000009 RLU18984.1 AXCN02001353 KY709722 AVK72273.1 AAZX01003032 AAZX01003033 AAZX01003034 AAZX01003035 AAZX01003041 AAZX01006162 AAZX01007365 AAZX01009310 AAZX01010627 AAZX01012590 CH477260 EAT45716.1 GDIP01163283 JAJ60119.1 KB031076 ELK03863.1 CM001291 EHH57720.1 HAAF01007547 CCP79371.1 GFDL01003606 JAV31439.1 GFDL01003607 JAV31438.1 JT410717 AHH39167.1 EAT45711.1 JW862400 AFO94917.1 JW862119 AFO94636.1 GCES01057181 JAR29142.1 CM004482 OCT63109.1 CP026250 AWP05836.1 AL928622 BX255899 AF002717 CU638812 AAC60252.1 CAP71961.1 KM655584 AJG05930.1 AAMC01064985 AAMC01064986 AAMC01064987 AAMC01064988 AAMC01064989 AAMC01064990 AAMC01064991 AJFE02061783 AJFE02061784 AJFE02061785 AJFE02061786 AJFE02061787 AJFE02061788 AJFE02061789 AJFE02061790 AJFE02061791 AJFE02061792 GABZ01001230 JAA52295.1 CH916370 EDV99782.1 CH902660 EDV30112.2 AY060671 AAL28219.1 AF033341 AAB87983.1 CP012528 ALC49294.1 JH882969 ELR47748.1 CH479196 EDW27672.1
KQ460210 KPJ16592.1 RSAL01000104 RVE47366.1 AGBW02008998 OWR51896.1 GEDC01020017 JAS17281.1 GECU01019739 GECU01014871 JAS87967.1 JAS92835.1 JTDY01000076 KOB79002.1 GEBQ01021277 JAT18700.1 ADTU01001922 ADTU01001923 ADTU01001924 ADTU01001925 ADTU01001926 ADTU01001927 ADTU01001928 ADTU01001929 ADTU01001930 ADTU01001931 KK852567 KDR21223.1 GBYB01001992 GBYB01001993 JAG71759.1 JAG71760.1 GEZM01042509 JAV79809.1 KQ435005 KZC13407.1 GECZ01026511 JAS43258.1 GL450619 EFN80728.1 GEZM01042511 JAV79807.1 KQ973114 EFA12173.2 GBHO01005368 JAG38236.1 DS235848 EEB18560.1 GEZM01042510 JAV79808.1 GDIQ01204138 JAK47587.1 GGMS01007042 MBY76245.1 GDHC01000349 JAQ18280.1 GDIP01240540 JAI82861.1 GDIQ01122073 JAL29653.1 GDIQ01021178 JAN73559.1 GDIP01250809 JAI72592.1 KQ435692 KOX81075.1 GGMR01011803 MBY24422.1 GFXV01003306 MBW15111.1 AXCM01009261 GL732577 EFX75164.1 LNIX01000004 OXA55423.1 AJWK01002192 AJWK01002193 AJWK01002194 APCN01001217 LJIJ01000678 ODM95397.1 CAEY01001560 GAMD01002071 JAA99519.1 QOIP01000009 RLU18984.1 AXCN02001353 KY709722 AVK72273.1 AAZX01003032 AAZX01003033 AAZX01003034 AAZX01003035 AAZX01003041 AAZX01006162 AAZX01007365 AAZX01009310 AAZX01010627 AAZX01012590 CH477260 EAT45716.1 GDIP01163283 JAJ60119.1 KB031076 ELK03863.1 CM001291 EHH57720.1 HAAF01007547 CCP79371.1 GFDL01003606 JAV31439.1 GFDL01003607 JAV31438.1 JT410717 AHH39167.1 EAT45711.1 JW862400 AFO94917.1 JW862119 AFO94636.1 GCES01057181 JAR29142.1 CM004482 OCT63109.1 CP026250 AWP05836.1 AL928622 BX255899 AF002717 CU638812 AAC60252.1 CAP71961.1 KM655584 AJG05930.1 AAMC01064985 AAMC01064986 AAMC01064987 AAMC01064988 AAMC01064989 AAMC01064990 AAMC01064991 AJFE02061783 AJFE02061784 AJFE02061785 AJFE02061786 AJFE02061787 AJFE02061788 AJFE02061789 AJFE02061790 AJFE02061791 AJFE02061792 GABZ01001230 JAA52295.1 CH916370 EDV99782.1 CH902660 EDV30112.2 AY060671 AAL28219.1 AF033341 AAB87983.1 CP012528 ALC49294.1 JH882969 ELR47748.1 CH479196 EDW27672.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000007151
UP000037510
+ More
UP000005205 UP000027135 UP000076502 UP000008237 UP000007266 UP000009046 UP000053105 UP000075883 UP000000305 UP000198287 UP000075885 UP000092461 UP000075840 UP000094527 UP000075880 UP000076407 UP000075903 UP000015104 UP000279307 UP000075886 UP000075920 UP000075900 UP000075884 UP000002358 UP000008820 UP000095301 UP000010552 UP000009130 UP000221080 UP000018467 UP000186698 UP000265040 UP000001646 UP000246464 UP000000437 UP000192224 UP000008143 UP000261360 UP000261420 UP000261480 UP000264820 UP000087266 UP000261400 UP000261500 UP000257160 UP000002280 UP000265140 UP000007635 UP000240080 UP000001070 UP000007801 UP000002852 UP000092553 UP000233140 UP000233220 UP000008744 UP000265080
UP000005205 UP000027135 UP000076502 UP000008237 UP000007266 UP000009046 UP000053105 UP000075883 UP000000305 UP000198287 UP000075885 UP000092461 UP000075840 UP000094527 UP000075880 UP000076407 UP000075903 UP000015104 UP000279307 UP000075886 UP000075920 UP000075900 UP000075884 UP000002358 UP000008820 UP000095301 UP000010552 UP000009130 UP000221080 UP000018467 UP000186698 UP000265040 UP000001646 UP000246464 UP000000437 UP000192224 UP000008143 UP000261360 UP000261420 UP000261480 UP000264820 UP000087266 UP000261400 UP000261500 UP000257160 UP000002280 UP000265140 UP000007635 UP000240080 UP000001070 UP000007801 UP000002852 UP000092553 UP000233140 UP000233220 UP000008744 UP000265080
Interpro
Gene 3D
ProteinModelPortal
H9JQX5
A0A2W1BZJ7
A0A194PV02
A0A0N1PHN9
A0A3S2TJ02
A0A212FDR4
+ More
A0A1B6CV77 A0A1B6J0X0 A0A0L7LU79 A0A1B6L4V9 A0A158NJF8 A0A067RDY5 A0A0C9PLN5 A0A1Y1M261 A0A154PNU8 A0A1B6EZB2 E2BUA0 A0A1Y1M907 D7EHQ2 A0A0A9Z2X5 E0VYV4 A0A1Y1M443 A0A0P5JJJ3 A0A2S2QES5 A0A146MHX1 A0A0P4XQH6 A0A0P5RCR6 A0A0P6HWB0 A0A0N7ZFE1 A0A0M9ABD1 A0A2S2P501 A0A2H8TME8 A0A182MGN3 E9GZG2 A0A226ECW7 A0A182PN48 A0A1B0C9C2 A0A182IAE8 A0A1D2MQP3 A0A182JHX5 A0A182WTN4 A0A182V2Z4 T1K3U5 T1DI33 A0A3L8DF10 A0A182QSJ5 A0A182WQJ7 A0A182S4K3 A0A2P1DV55 A0A182NZ13 K7J7Q0 Q0IGC3 A0A1I8N3U9 A0A0P5DCN6 L5K039 G7PTM1 U6D9U2 A0A1Q3FV05 A0A1Q3FV72 W5UHM1 Q0IGC8 V9KAW1 W5KQW5 V9K9V4 A0A146WJ74 A0A1L8EV13 A0A3Q1INQ5 G1KUB3 A0A2U9BQD3 O42140 A0A0B5JPZ7 A0A1W5A9T8 F7CBX5 A0A3B4WFQ0 A0A3B4VCM8 A0A3B3XTX2 A0A3Q2YN80 A0A1S3PS44 A0A3B5B6R7 A0A3B3W2Z4 A0A3Q1B3Y3 F7G4D6 A0A3P8XKC4 G3Q6U6 A0A2R9AA25 K9ITB3 B4JJN0 B3N1K3 Q95SP5 O57339 M3ZWP7 A0A0M5JDR0 L8HV67 A0A2K5ZVT1 A0A2K6SXG2 B4GY45 A0A3B1KE85 A0A1S3L5W0 A0A3P8RME6
A0A1B6CV77 A0A1B6J0X0 A0A0L7LU79 A0A1B6L4V9 A0A158NJF8 A0A067RDY5 A0A0C9PLN5 A0A1Y1M261 A0A154PNU8 A0A1B6EZB2 E2BUA0 A0A1Y1M907 D7EHQ2 A0A0A9Z2X5 E0VYV4 A0A1Y1M443 A0A0P5JJJ3 A0A2S2QES5 A0A146MHX1 A0A0P4XQH6 A0A0P5RCR6 A0A0P6HWB0 A0A0N7ZFE1 A0A0M9ABD1 A0A2S2P501 A0A2H8TME8 A0A182MGN3 E9GZG2 A0A226ECW7 A0A182PN48 A0A1B0C9C2 A0A182IAE8 A0A1D2MQP3 A0A182JHX5 A0A182WTN4 A0A182V2Z4 T1K3U5 T1DI33 A0A3L8DF10 A0A182QSJ5 A0A182WQJ7 A0A182S4K3 A0A2P1DV55 A0A182NZ13 K7J7Q0 Q0IGC3 A0A1I8N3U9 A0A0P5DCN6 L5K039 G7PTM1 U6D9U2 A0A1Q3FV05 A0A1Q3FV72 W5UHM1 Q0IGC8 V9KAW1 W5KQW5 V9K9V4 A0A146WJ74 A0A1L8EV13 A0A3Q1INQ5 G1KUB3 A0A2U9BQD3 O42140 A0A0B5JPZ7 A0A1W5A9T8 F7CBX5 A0A3B4WFQ0 A0A3B4VCM8 A0A3B3XTX2 A0A3Q2YN80 A0A1S3PS44 A0A3B5B6R7 A0A3B3W2Z4 A0A3Q1B3Y3 F7G4D6 A0A3P8XKC4 G3Q6U6 A0A2R9AA25 K9ITB3 B4JJN0 B3N1K3 Q95SP5 O57339 M3ZWP7 A0A0M5JDR0 L8HV67 A0A2K5ZVT1 A0A2K6SXG2 B4GY45 A0A3B1KE85 A0A1S3L5W0 A0A3P8RME6
PDB
4PLO
E-value=1.53492e-54,
Score=536
Ontologies
GO
GO:0005576
GO:0008045
GO:0005604
GO:0009887
GO:0016358
GO:0009888
GO:0016021
GO:0001764
GO:0007097
GO:0030334
GO:0061643
GO:0030517
GO:0008284
GO:0005737
GO:0051963
GO:0098609
GO:0033564
GO:0045773
GO:0042472
GO:0001944
GO:0001947
GO:0071678
GO:0001569
GO:0016525
GO:0001525
GO:0048665
GO:0048846
GO:0021952
GO:0030516
GO:0030878
GO:0021960
GO:0061626
GO:0003146
GO:0035908
GO:0060603
GO:0005515
GO:0005615
GO:0008083
GO:0005179
GO:0015074
GO:0015923
GO:0005634
GO:0006260
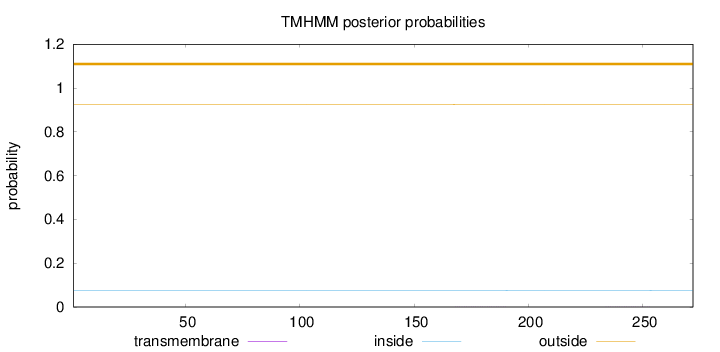
Topology
Subcellular location
Secreted
Length:
272
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01185
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07590
outside
1 - 272
Population Genetic Test Statistics
Pi
242.602632
Theta
173.167844
Tajima's D
1.280829
CLR
0.377672
CSRT
0.735463226838658
Interpretation
Uncertain
