Gene
KWMTBOMO06392
Pre Gene Modal
BGIBMGA011929
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_K02A2.6-like_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 1.66
Sequence
CDS
ATGAAGATTCTCCTTGCTATTGCATTAATATTGTCAACAGTAATGTGGGTGTCAACACAACAGCCACAAGGAGTGCACACGTACTGCGGGCGCCACTTGGCTCGCACTCTGGCCGACCTGTGTTGGGAAGCGGGCGTGGACAAGCGCAGCGGTGCTCAGTTCGCGAGCTACGGCTCCGCGTGGCTGATGCCGTACTCGGAAGGACGCGGCAAACGAGGCATCGTGGATGAGTGCTGTCTCAGACCCTGCAGCGTGGACGTGCTTCTGTCCATGCAAACTTGGAAGACTTGGAGATGCAGACTCACACAGTGGTTCATCGCGAACGATATTAACGTGGTGAAAGATCCAGCAGGAGTCAAAAGACGGGCCATATTGCTTAGCACACTCAGCGAATCTACCTACAAACTGGCAGCTGATCTCGTGCTGCCTAAAGAAGTTCAAGAAGTACCATACGAAGATATAGTTAGTGCCTTGGACAACCATTTCACTCCAAAATCAGTGGGATTTGGCGAGCGTCATCATTTTTATGCCGCCACACAACTACCCCAGGAGACACCATCACAGTGGGCTGCGAGGTTACGAGGTCTCACTTCTCAGTGTAACTTCAGCAATGTAGAAGAAGCATTAAGAGATCGATTCATTATGGGTATGCTGCCGGGCGTGGAGAAAGAAAAAATGTACGTGCAGGAGCTGGCCGGGCTGACCTTCGCCAGGGCTGTGGAGCTGACCGAGAGCTTGCGCAGCGCCCGCGCGGCTGCTGCTGCAAGCGCTGCTGCCGAGACTGTCGTTACAGAGCTGTACAAAATAGGCAAACGTTCTCCGACTAGTGTACCTCAAAGTGCAAAAGTAAAGTGTTCCGTGTGTGGTTATTCGAATCACAAAGCCGCGGAGTGTCGTTTTGCGAGTTACACTTGTAGGAAGTGCAACAAAAAAGGACATTTGCGTAGAATGTGCAAAAGAATCAATTATGTTGTAACGGAGGAGAACGATGAAGGTGACGGTGACGACGGTAAAATTTGTAACATTCGATCTTTGCGAGGTGAACCATTAATCGAAGCCGTGTGTGTCAATGGTATCAGAATGAACTTTGAGATTGATAGTGGGTCAGCTGTTACCGTTATGTCCGTAGATACATATAACGAACATTTTAAAAACATTCCCTTATTTAGCACAAATAAGAAGTTATTAAGTTACTCGGGTGACGTGTTAGGTTGCGCCGGTCGCGCTCAGTTGTTAGTGGAGTGGCGGGGTCGCACGCGCCAGCTCGACGTGTACGTGGTGCGCGATGGCGGACCGCCTTTACTGGGCCGCGACTTCATCGCACAATTTGAACTGCAGCTAGCGCCCGTATATAAATGTGCCACTACCCGTGACACGTTGAAATTAATACAGGAACAATATCCCGCCGTGTTCTCGGACAAGCTAGGTAAATTCAACAAATATAAAGTTAATTTGAAATTAAAAGAAAGTGCTCATCCTATATTTTTTAAACCACGGCCGGTCGCATTTGCTCTGCGTGAAAAAGTGGAAAAAGAGATAAACCGTTTGGTGGGACTGGGAGTACTCCGACCAGTCGAGCATTCCGACTACGCGTCACCAATAGTCCCGGTACTGAAACGTGACGGTTCGGTCCGTATCTGCGCGGACTACTCTGTTACTATAAACAAGCAATTAGTTGTAGAACAGTATCCGTTGCCTACCGTAAACGAGTTGTTTTCTAAACTATACGGGGGCCAGAAATTTACTAAATTAGATTTGTCTATGGCCTACGGTCAGTTTTGTCTGGATGAAGAATCCCAAAAGTTAACTTGTATCAATACGCATAAAGGGATCTTCGCGTATACGCGTTTGGTGTTCGGGTTAGCTAGTGGGCCATCAATTTTCCAGCGAGCCATGGATACAGTGCTAGCGGGCCTGGACGGCACGCTGTGTTTATTGGACGATGTGCTCATAACGGGTCGCAATGACTCTGAACATCTTGAGAGATTGCATCAGGTCCTAAAACGGTTGCAAGACGCAGGATTTGTTCTACAAAAGTCTAAATCTACGACCGACTCCGCCGCGCACTCCGGTGTGCCCGTGGGAATACCCCCACAACCCTTTTACCGTGTACACTTAGACTTTCTAGGTCCGCTGAATGGTCGGATGTACCTGGTCCTAGTAGACGCGTACTCCAAGTGGCTTGAGGTGTACGAGATGGCTTCCACAACTACTAGTGCGGTCATTGAAAGAATGTATGAGTACATGTCCAGATACGGATTGCCTCATACGGTAGTTACGGATAACGGTGTGATATTAAAGAGAATTGGTAAAAACGTATACATTGTAAAAGACTTTAAATCTTCAATACAACTCAAGAGGCACTCTGAACAGCTGTTTTCGTGCAAAACCAAATGTGGTGCCAGTCAAACCTGGGCGGATGACTTGAGCGGTTCTATTGCTCCAACCGTTGCTAGTCCGGCAACGGGGATGATGTCGTCGCCTTGTGATGATAGTAGCGGCATCGGACCTCACACACACGACGAAGAGAGTGCAACTGAGGAGAGGGAAGAGGAGAGCGGCCACACTGGAATCTCTAGTGCATGTGACAATCAGCCGAATCGGGAGTCACCGGAACCGAGTCACGTAACTGTCACGCGAGGAGTGCGTCGACCCGCGCAGTCTGAAGACTCTGAAGAAGGGGATCTGTTCTTCGAGGCAGAGGAGGTGAGCTTGGAGCCCCAAAATATTAGGGAGCCCAGGTTTCCTAAACGTAACCGTCCTAGGGTTAATTATAGACAGTTTTATTAA
Protein
MKILLAIALILSTVMWVSTQQPQGVHTYCGRHLARTLADLCWEAGVDKRSGAQFASYGSAWLMPYSEGRGKRGIVDECCLRPCSVDVLLSMQTWKTWRCRLTQWFIANDINVVKDPAGVKRRAILLSTLSESTYKLAADLVLPKEVQEVPYEDIVSALDNHFTPKSVGFGERHHFYAATQLPQETPSQWAARLRGLTSQCNFSNVEEALRDRFIMGMLPGVEKEKMYVQELAGLTFARAVELTESLRSARAAAAASAAAETVVTELYKIGKRSPTSVPQSAKVKCSVCGYSNHKAAECRFASYTCRKCNKKGHLRRMCKRINYVVTEENDEGDGDDGKICNIRSLRGEPLIEAVCVNGIRMNFEIDSGSAVTVMSVDTYNEHFKNIPLFSTNKKLLSYSGDVLGCAGRAQLLVEWRGRTRQLDVYVVRDGGPPLLGRDFIAQFELQLAPVYKCATTRDTLKLIQEQYPAVFSDKLGKFNKYKVNLKLKESAHPIFFKPRPVAFALREKVEKEINRLVGLGVLRPVEHSDYASPIVPVLKRDGSVRICADYSVTINKQLVVEQYPLPTVNELFSKLYGGQKFTKLDLSMAYGQFCLDEESQKLTCINTHKGIFAYTRLVFGLASGPSIFQRAMDTVLAGLDGTLCLLDDVLITGRNDSEHLERLHQVLKRLQDAGFVLQKSKSTTDSAAHSGVPVGIPPQPFYRVHLDFLGPLNGRMYLVLVDAYSKWLEVYEMASTTTSAVIERMYEYMSRYGLPHTVVTDNGVILKRIGKNVYIVKDFKSSIQLKRHSEQLFSCKTKCGASQTWADDLSGSIAPTVASPATGMMSSPCDDSSGIGPHTHDEESATEEREEESGHTGISSACDNQPNRESPEPSHVTVTRGVRRPAQSEDSEEGDLFFEAEEVSLEPQNIREPRFPKRNRPRVNYRQFY
Summary
Uniprot
EMBL
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR021109 Peptidase_aspartic_dom_sf
IPR036875 Znf_CCHC_sf
IPR041577 RT_RNaseH_2
IPR041588 Integrase_H2C2
IPR001995 Peptidase_A2_cat
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR036397 RNaseH_sf
IPR001878 Znf_CCHC
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR021109 Peptidase_aspartic_dom_sf
IPR036875 Znf_CCHC_sf
IPR041577 RT_RNaseH_2
IPR041588 Integrase_H2C2
IPR001995 Peptidase_A2_cat
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR036397 RNaseH_sf
IPR001878 Znf_CCHC
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR001969 Aspartic_peptidase_AS
Gene 3D
ProteinModelPortal
PDB
4OL8
E-value=4.40735e-24,
Score=279
Ontologies
GO
Topology
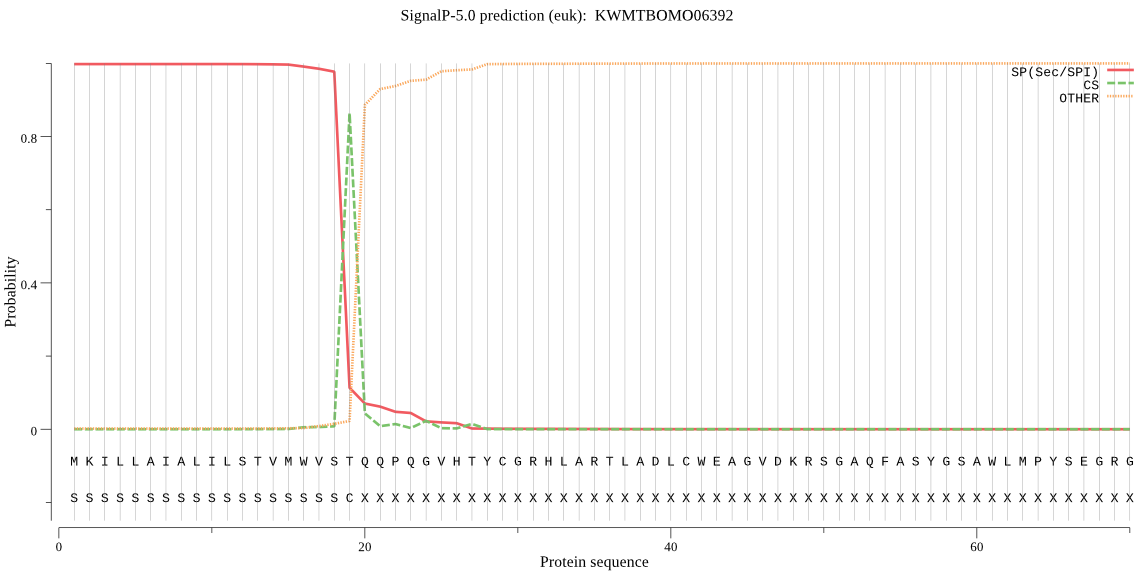
SignalP
Position: 1 - 19,
Likelihood: 0.997922
Length:
929
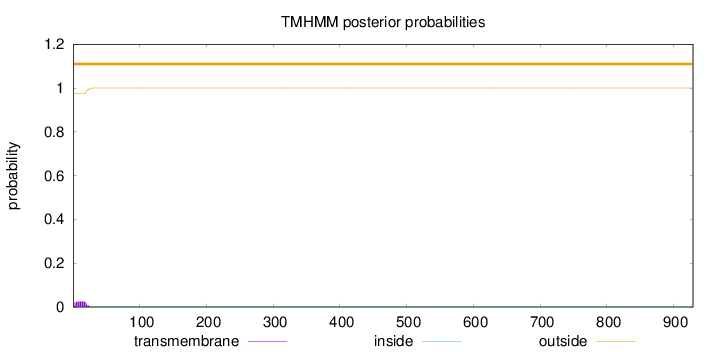
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.47014
Exp number, first 60 AAs:
0.46665
Total prob of N-in:
0.02455
outside
1 - 929
Population Genetic Test Statistics
Pi
231.318397
Theta
176.962094
Tajima's D
1.115792
CLR
47.753828
CSRT
0.693965301734913
Interpretation
Uncertain
