Gene
KWMTBOMO06383
Pre Gene Modal
BGIBMGA011925
Annotation
PREDICTED:_netrin-B_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.618
Sequence
CDS
ATGGCAGCTCTGGTGTCGGCGCTGCTGCTGGCAGTGTTGCTGGCTGCCGCGGTGGCCACCCCACCGGACCCCTGCTACGAGGAGTCGCGCCCTCGGAGGTGCATCCCCGATTTCGTGAATGCTGCTTTCGGTGCCCCCGTGATCGCGTCTTCAGCGTGCGGCCGTCGCGGGCCCGAGAACCTGTGCGACCCTCCACTCGACCAGTCTCCGGAGCTGGCTTGCAGGGTGTGCGATGCGGCTCGGCCTGACCGCCGCCACCCGTCGCTTTATCTCACAGACCTCAACAACCCCAATAACATAACGTGTTGGCGCTCGGCCGCGCTCATGCCAGCCTCGTACGACTCACCTCCCGACAATGTGTCGCTGACCCTCTCCCTCGGGAAGAAATATGAGTTGACGTACATCAGCCTACAGTTCTGTCCGAAGTCTGCCCGACCAGACTCAGTGGCGATCTACAAGAGTGCAGATTACGGGCTCAACTGGCAACCTTTCCAGTTCTACTCGAGTCAGTGTCGGCGGGTGTACGGCCGGCCCAACAAGGCCACGGTGACCGCCGCGAACGAGCAGGAGGCGAGGTGCTCCGACACCCACCGTCTGGCCGGCGACAGTGCAGGAGGCGCGCGACTTGCATTTAGTACCCTAGAGGGTCGACCCAGCGCGGCCAACTTCGACCTCTCCCCTGTGCTTCAGGACTGGGTGACCGCGACAGACGTGAGAGTCGTGTTTAATCGCTTGCACATGGTCAACGAAGGCGACGAGATAGATTTGAAGAAGCAGGCGGCTCCTGGCACACAGCACTACGCGGTATCTGATTTCGCCGTCGGAGGTCGGTGCAAGTGCAACGGGCACGCGTCGCGCTGCATCCCGATGAAGGAAGGAGAGGGCTCCAGCGCTGGTGGAGTAGTGCAACTGGCCTGCGACTGCAAGCACAACACCGCCGGTCGAGATTGCGAGAGATGCAAGCCGTTCCACTTCGATCGACCCTGGGGCCGCGCGACCGCCCGAGACGCCAACGAATGTAAAGGTCAGTCTAATGTTTATCACGGAATCGGAAAAATGTAG
Protein
MAALVSALLLAVLLAAAVATPPDPCYEESRPRRCIPDFVNAAFGAPVIASSACGRRGPENLCDPPLDQSPELACRVCDAARPDRRHPSLYLTDLNNPNNITCWRSAALMPASYDSPPDNVSLTLSLGKKYELTYISLQFCPKSARPDSVAIYKSADYGLNWQPFQFYSSQCRRVYGRPNKATVTAANEQEARCSDTHRLAGDSAGGARLAFSTLEGRPSAANFDLSPVLQDWVTATDVRVVFNRLHMVNEGDEIDLKKQAAPGTQHYAVSDFAVGGRCKCNGHASRCIPMKEGEGSSAGGVVQLACDCKHNTAGRDCERCKPFHFDRPWGRATARDANECKGQSNVYHGIGKM
Summary
Uniprot
H9JQW9
A0A2A4JNT5
A0A2W1C0P9
A0A2H1VCB8
A0A194PUR7
A0A0N0PF41
+ More
A0A0L7LH47 A0A195B5K7 A0A0L7QW80 A0A026W9I1 T1I1I5 A0A310SFK5 A0A0M9ABD9 A0A195FQB3 A0A1B0DHY6 B0XC85 A0A154PNG0 A0A1B0CDE2 K7J7Q0 Q0IGC8 Q0IGC2 B0XFM2 T1IHF5 A0A2P6KWY7 A0A1Q3FV05 A0A1Q3FV72 A0A336MLD0 A0A087TQ16 A0A336M6K0 B7P5R8 V4AN98 A0A3S3RY09 A0A336MIN0 A0A0P7TYR9 F5BU38 A0A2P1DV55 A0A0P7XL33 A0A165USS5 Q9NFW6 A0A060XZ03 A0A060W5M7 A0A0P5W134 A0A0P6HRZ8 A0A0P4XYZ6 A0A0N8CYF0 A0A1J1I943 A0A2I4AY51 A0A3B1KAR5 A0A3N0XFW4 A0A0N8D0A7 A0A0P5AIN2 A0A0P6EJ88 A0A3B4BS53 T1II68 A0A060X0C3 A0A060WRZ6 A0A0P5NGS2 A0A3N0YAZ9 A0A3B4BNG7 A0A2I4AY75 Q29RD2 A0A3B1JST2 A0A0R4IN40 A0A2I4AY73 A0A2I4AY69 A0A0P4ZHD4 A0A0P6G559 A6YB92 A0A164U1Q7 W5N6U6 A0A3B5LRA3 A0A315WA52 A0A3P8XKA9 A0A0P5HX01 A0A0N7ZRE0 A0A0P4WD43 A0A3R7SKF6 A0A2P4SS64 A0A3Q1F2U3 A0A1S3JF50 A0A1S3JED4 A0A3B3CFV1 A0A1S3JDN1 A0A3P8XKC4 A0A1S3JDW4 A0A1S3JDP2 A0A3B3ZZV3 A0A0P5RCR6 A0A3B5M0C3 A0A3B4GE39 A0A147A3Q7 A0A3Q1JLT0 A0A3Q2UXU7 A0A3Q3GV46 A0A3B3YIJ3 A0A3Q2Z716 A0A0B5JK74 A0A3Q2YSH1 A0A3B4ZCK8 A0A3B4WH08
A0A0L7LH47 A0A195B5K7 A0A0L7QW80 A0A026W9I1 T1I1I5 A0A310SFK5 A0A0M9ABD9 A0A195FQB3 A0A1B0DHY6 B0XC85 A0A154PNG0 A0A1B0CDE2 K7J7Q0 Q0IGC8 Q0IGC2 B0XFM2 T1IHF5 A0A2P6KWY7 A0A1Q3FV05 A0A1Q3FV72 A0A336MLD0 A0A087TQ16 A0A336M6K0 B7P5R8 V4AN98 A0A3S3RY09 A0A336MIN0 A0A0P7TYR9 F5BU38 A0A2P1DV55 A0A0P7XL33 A0A165USS5 Q9NFW6 A0A060XZ03 A0A060W5M7 A0A0P5W134 A0A0P6HRZ8 A0A0P4XYZ6 A0A0N8CYF0 A0A1J1I943 A0A2I4AY51 A0A3B1KAR5 A0A3N0XFW4 A0A0N8D0A7 A0A0P5AIN2 A0A0P6EJ88 A0A3B4BS53 T1II68 A0A060X0C3 A0A060WRZ6 A0A0P5NGS2 A0A3N0YAZ9 A0A3B4BNG7 A0A2I4AY75 Q29RD2 A0A3B1JST2 A0A0R4IN40 A0A2I4AY73 A0A2I4AY69 A0A0P4ZHD4 A0A0P6G559 A6YB92 A0A164U1Q7 W5N6U6 A0A3B5LRA3 A0A315WA52 A0A3P8XKA9 A0A0P5HX01 A0A0N7ZRE0 A0A0P4WD43 A0A3R7SKF6 A0A2P4SS64 A0A3Q1F2U3 A0A1S3JF50 A0A1S3JED4 A0A3B3CFV1 A0A1S3JDN1 A0A3P8XKC4 A0A1S3JDW4 A0A1S3JDP2 A0A3B3ZZV3 A0A0P5RCR6 A0A3B5M0C3 A0A3B4GE39 A0A147A3Q7 A0A3Q1JLT0 A0A3Q2UXU7 A0A3Q3GV46 A0A3B3YIJ3 A0A3Q2Z716 A0A0B5JK74 A0A3Q2YSH1 A0A3B4ZCK8 A0A3B4WH08
Pubmed
EMBL
BABH01028601
NWSH01001020
PCG73080.1
KZ149897
PZC78690.1
ODYU01001780
+ More
SOQ38455.1 KQ459590 KPI97151.1 KQ459692 KPJ20422.1 JTDY01001164 KOB74684.1 KQ976582 KYM79798.1 KQ414716 KOC62857.1 KK107320 EZA52767.1 ACPB03016596 KQ761843 OAD56602.1 KQ435692 KOX81077.1 KQ981305 KYN42780.1 AJVK01006076 AJVK01006077 AJVK01006078 DS232678 EDS44691.1 KQ435005 KZC13406.1 AJWK01007724 AAZX01003032 AAZX01003033 AAZX01003034 AAZX01003035 AAZX01003041 AAZX01006162 AAZX01007365 AAZX01009310 AAZX01010627 AAZX01012590 CH477260 EAT45711.1 EAT45717.1 DS232941 EDS26885.1 JH429903 MWRG01004066 PRD30845.1 GFDL01003606 JAV31439.1 GFDL01003607 JAV31438.1 UFQS01001396 UFQT01001396 SSX10679.1 SSX30361.1 KK116254 KFM67205.1 UFQT01000629 SSX25886.1 ABJB010969459 DS642248 EEC01940.1 KB201234 ESO98632.1 NCKU01003340 RWS07715.1 SSX25888.1 JARO02008327 KPP62856.1 JF302896 ADY05326.1 KY709722 AVK72273.1 JARO02001333 KPP75780.1 KR232551 AMY99575.1 AJ252166 CAB72422.1 FR906523 CDQ84691.1 FR904335 CDQ59855.1 GDIP01093481 JAM10234.1 GDIQ01039810 JAN54927.1 GDIP01234933 JAI88468.1 GDIP01086647 JAM17068.1 CVRI01000044 CRK96739.1 RJVU01079141 ROI15691.1 GDIP01081391 JAM22324.1 GDIP01198518 JAJ24884.1 GDIQ01061994 JAN32743.1 JH430143 FR904733 CDQ70739.1 FR904598 CDQ67834.1 GDIQ01145524 JAL06202.1 RJVU01049122 ROL42928.1 BC114259 AAI14260.1 BX255899 GDIP01226071 JAI97330.1 GDIQ01039809 JAN54928.1 EF384215 ABO93214.1 LRGB01001636 KZS10976.1 AHAT01000424 AHAT01000425 NHOQ01000158 PWA32634.1 GDIQ01223785 JAK27940.1 GDIP01220017 JAJ03385.1 GDRN01048943 JAI66631.1 QCYY01003210 ROT64683.1 PPHD01026137 POI26937.1 GDIQ01122073 JAL29653.1 GCES01013194 JAR73129.1 KM655683 AJG06029.1
SOQ38455.1 KQ459590 KPI97151.1 KQ459692 KPJ20422.1 JTDY01001164 KOB74684.1 KQ976582 KYM79798.1 KQ414716 KOC62857.1 KK107320 EZA52767.1 ACPB03016596 KQ761843 OAD56602.1 KQ435692 KOX81077.1 KQ981305 KYN42780.1 AJVK01006076 AJVK01006077 AJVK01006078 DS232678 EDS44691.1 KQ435005 KZC13406.1 AJWK01007724 AAZX01003032 AAZX01003033 AAZX01003034 AAZX01003035 AAZX01003041 AAZX01006162 AAZX01007365 AAZX01009310 AAZX01010627 AAZX01012590 CH477260 EAT45711.1 EAT45717.1 DS232941 EDS26885.1 JH429903 MWRG01004066 PRD30845.1 GFDL01003606 JAV31439.1 GFDL01003607 JAV31438.1 UFQS01001396 UFQT01001396 SSX10679.1 SSX30361.1 KK116254 KFM67205.1 UFQT01000629 SSX25886.1 ABJB010969459 DS642248 EEC01940.1 KB201234 ESO98632.1 NCKU01003340 RWS07715.1 SSX25888.1 JARO02008327 KPP62856.1 JF302896 ADY05326.1 KY709722 AVK72273.1 JARO02001333 KPP75780.1 KR232551 AMY99575.1 AJ252166 CAB72422.1 FR906523 CDQ84691.1 FR904335 CDQ59855.1 GDIP01093481 JAM10234.1 GDIQ01039810 JAN54927.1 GDIP01234933 JAI88468.1 GDIP01086647 JAM17068.1 CVRI01000044 CRK96739.1 RJVU01079141 ROI15691.1 GDIP01081391 JAM22324.1 GDIP01198518 JAJ24884.1 GDIQ01061994 JAN32743.1 JH430143 FR904733 CDQ70739.1 FR904598 CDQ67834.1 GDIQ01145524 JAL06202.1 RJVU01049122 ROL42928.1 BC114259 AAI14260.1 BX255899 GDIP01226071 JAI97330.1 GDIQ01039809 JAN54928.1 EF384215 ABO93214.1 LRGB01001636 KZS10976.1 AHAT01000424 AHAT01000425 NHOQ01000158 PWA32634.1 GDIQ01223785 JAK27940.1 GDIP01220017 JAJ03385.1 GDRN01048943 JAI66631.1 QCYY01003210 ROT64683.1 PPHD01026137 POI26937.1 GDIQ01122073 JAL29653.1 GCES01013194 JAR73129.1 KM655683 AJG06029.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000078540
+ More
UP000053825 UP000053097 UP000015103 UP000053105 UP000078541 UP000092462 UP000002320 UP000076502 UP000092461 UP000002358 UP000008820 UP000054359 UP000001555 UP000030746 UP000285301 UP000034805 UP000193380 UP000183832 UP000192220 UP000018467 UP000261440 UP000000437 UP000076858 UP000018468 UP000261380 UP000265140 UP000283509 UP000257200 UP000085678 UP000261560 UP000261520 UP000261460 UP000265040 UP000264840 UP000261660 UP000261480 UP000264820 UP000261400 UP000261360
UP000053825 UP000053097 UP000015103 UP000053105 UP000078541 UP000092462 UP000002320 UP000076502 UP000092461 UP000002358 UP000008820 UP000054359 UP000001555 UP000030746 UP000285301 UP000034805 UP000193380 UP000183832 UP000192220 UP000018467 UP000261440 UP000000437 UP000076858 UP000018468 UP000261380 UP000265140 UP000283509 UP000257200 UP000085678 UP000261560 UP000261520 UP000261460 UP000265040 UP000264840 UP000261660 UP000261480 UP000264820 UP000261400 UP000261360
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JQW9
A0A2A4JNT5
A0A2W1C0P9
A0A2H1VCB8
A0A194PUR7
A0A0N0PF41
+ More
A0A0L7LH47 A0A195B5K7 A0A0L7QW80 A0A026W9I1 T1I1I5 A0A310SFK5 A0A0M9ABD9 A0A195FQB3 A0A1B0DHY6 B0XC85 A0A154PNG0 A0A1B0CDE2 K7J7Q0 Q0IGC8 Q0IGC2 B0XFM2 T1IHF5 A0A2P6KWY7 A0A1Q3FV05 A0A1Q3FV72 A0A336MLD0 A0A087TQ16 A0A336M6K0 B7P5R8 V4AN98 A0A3S3RY09 A0A336MIN0 A0A0P7TYR9 F5BU38 A0A2P1DV55 A0A0P7XL33 A0A165USS5 Q9NFW6 A0A060XZ03 A0A060W5M7 A0A0P5W134 A0A0P6HRZ8 A0A0P4XYZ6 A0A0N8CYF0 A0A1J1I943 A0A2I4AY51 A0A3B1KAR5 A0A3N0XFW4 A0A0N8D0A7 A0A0P5AIN2 A0A0P6EJ88 A0A3B4BS53 T1II68 A0A060X0C3 A0A060WRZ6 A0A0P5NGS2 A0A3N0YAZ9 A0A3B4BNG7 A0A2I4AY75 Q29RD2 A0A3B1JST2 A0A0R4IN40 A0A2I4AY73 A0A2I4AY69 A0A0P4ZHD4 A0A0P6G559 A6YB92 A0A164U1Q7 W5N6U6 A0A3B5LRA3 A0A315WA52 A0A3P8XKA9 A0A0P5HX01 A0A0N7ZRE0 A0A0P4WD43 A0A3R7SKF6 A0A2P4SS64 A0A3Q1F2U3 A0A1S3JF50 A0A1S3JED4 A0A3B3CFV1 A0A1S3JDN1 A0A3P8XKC4 A0A1S3JDW4 A0A1S3JDP2 A0A3B3ZZV3 A0A0P5RCR6 A0A3B5M0C3 A0A3B4GE39 A0A147A3Q7 A0A3Q1JLT0 A0A3Q2UXU7 A0A3Q3GV46 A0A3B3YIJ3 A0A3Q2Z716 A0A0B5JK74 A0A3Q2YSH1 A0A3B4ZCK8 A0A3B4WH08
A0A0L7LH47 A0A195B5K7 A0A0L7QW80 A0A026W9I1 T1I1I5 A0A310SFK5 A0A0M9ABD9 A0A195FQB3 A0A1B0DHY6 B0XC85 A0A154PNG0 A0A1B0CDE2 K7J7Q0 Q0IGC8 Q0IGC2 B0XFM2 T1IHF5 A0A2P6KWY7 A0A1Q3FV05 A0A1Q3FV72 A0A336MLD0 A0A087TQ16 A0A336M6K0 B7P5R8 V4AN98 A0A3S3RY09 A0A336MIN0 A0A0P7TYR9 F5BU38 A0A2P1DV55 A0A0P7XL33 A0A165USS5 Q9NFW6 A0A060XZ03 A0A060W5M7 A0A0P5W134 A0A0P6HRZ8 A0A0P4XYZ6 A0A0N8CYF0 A0A1J1I943 A0A2I4AY51 A0A3B1KAR5 A0A3N0XFW4 A0A0N8D0A7 A0A0P5AIN2 A0A0P6EJ88 A0A3B4BS53 T1II68 A0A060X0C3 A0A060WRZ6 A0A0P5NGS2 A0A3N0YAZ9 A0A3B4BNG7 A0A2I4AY75 Q29RD2 A0A3B1JST2 A0A0R4IN40 A0A2I4AY73 A0A2I4AY69 A0A0P4ZHD4 A0A0P6G559 A6YB92 A0A164U1Q7 W5N6U6 A0A3B5LRA3 A0A315WA52 A0A3P8XKA9 A0A0P5HX01 A0A0N7ZRE0 A0A0P4WD43 A0A3R7SKF6 A0A2P4SS64 A0A3Q1F2U3 A0A1S3JF50 A0A1S3JED4 A0A3B3CFV1 A0A1S3JDN1 A0A3P8XKC4 A0A1S3JDW4 A0A1S3JDP2 A0A3B3ZZV3 A0A0P5RCR6 A0A3B5M0C3 A0A3B4GE39 A0A147A3Q7 A0A3Q1JLT0 A0A3Q2UXU7 A0A3Q3GV46 A0A3B3YIJ3 A0A3Q2Z716 A0A0B5JK74 A0A3Q2YSH1 A0A3B4ZCK8 A0A3B4WH08
PDB
4PLO
E-value=6.54773e-93,
Score=869
Ontologies
GO
Topology
Subcellular location
Secreted
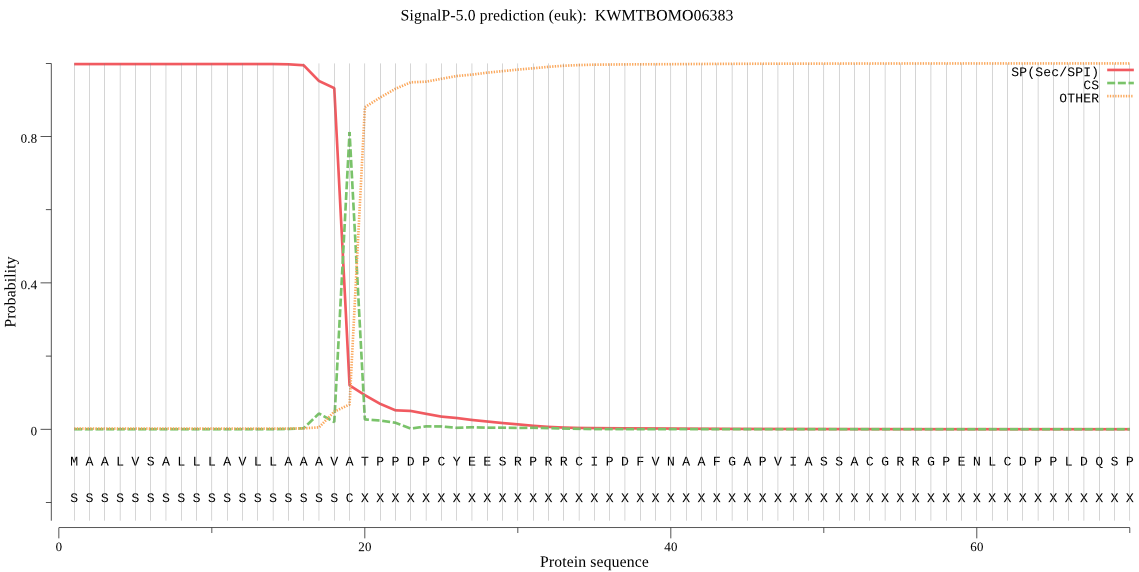
SignalP
Position: 1 - 19,
Likelihood: 0.997984
Length:
353
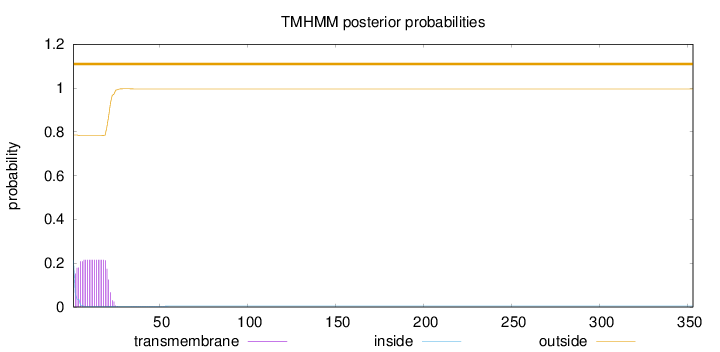
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.17367
Exp number, first 60 AAs:
4.17367
Total prob of N-in:
0.21449
outside
1 - 353
Population Genetic Test Statistics
Pi
330.895591
Theta
213.235311
Tajima's D
1.621617
CLR
0
CSRT
0.813409329533523
Interpretation
Uncertain
