Pre Gene Modal
BGIBMGA011973
Annotation
PREDICTED:_diphosphoinositol_polyphosphate_phosphohydrolase_1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.984
Sequence
CDS
ATGGTAAAGGAGAAGCCTAATTCGATAAGGATATATGACGACGAGGGTTTTAGGCGACGGGCGGCATGTATTTGCGTGCGTTCAGATGCCGAAACAGAGGTCCTGCTGGTGACGTCTTCGAGACGGCCCGACAACTGGATTGTGCCGGGCGGGGGAGTAGAGCCGGAAGAGGAGCCTTCTGTCACTGCGATGCGGGAGGTACTTGAGGAAGCCGGAGTGATCGGGAAACTAGGACGATGTCTTGGAGTATTCGAGAACCGAGAACACAAACACCGAACAGAGGTTTATGTTATGACTGTGACCCAAGAACTACCCGAGTGGGAGGATTCAAGACTCATGGGACGTAAGAGGCAATGGTTCTCCATTGATGATGCCCTGGCTCAGCTCGCCCTTCACAAGCCAATCCAGCGTCACTACCTTCAACAACTGCGTCGTTCCAAACAGAACAAGCAAGATGACCAGACTACGTATAACCTTCCCAGTTAG
Protein
MVKEKPNSIRIYDDEGFRRRAACICVRSDAETEVLLVTSSRRPDNWIVPGGGVEPEEEPSVTAMREVLEEAGVIGKLGRCLGVFENREHKHRTEVYVMTVTQELPEWEDSRLMGRKRQWFSIDDALAQLALHKPIQRHYLQQLRRSKQNKQDDQTTYNLPS
Summary
Similarity
Belongs to the Nudix hydrolase family.
Uniprot
H9JR13
A0A2H1VXY9
A0A2W1C3F6
A0A2A4JLZ0
A0A1E1WPC9
A0A212EJ29
+ More
A0A0N1IH85 A0A194Q176 A0A0L7L6P7 A0A0A1XQH5 A0A2W1BMN0 W8B688 A0A0K8WHD0 A0A034VTR0 A0A0K8TR98 T1PDT3 A0A1A9W4M9 D6WDJ7 U5EVU2 A0A0L0C1T9 A0A1Y1L4X1 T1DIN2 D3TM32 T1E2Y0 A0A1L8DLT2 A0A1W4XMX2 A0A1B0G5R0 A0A1A9ZMT9 A0A146M500 A0A1Q3EU50 A0A1S4JVA9 A0A182INR5 A0A1Q3EU45 A0A182M0R3 A0A1Q3EU35 A0A182GB38 A0A182RR76 A0A1I8PP03 A0A182U3C1 A0A182NAR9 A0A182UM49 A0A2C9GR99 A0A182KTK5 Q7Q196 A0A182XJK9 A0A182Q1R7 A0A182KGK4 A0A2M4BXZ3 A0A182F3V4 W5JW34 A0A2M4ANY6 A0A2M4AN06 A0A2M4ANQ2 A0A2M3ZI64 A0A1B0CHT3 Q17EU8 A0A2M3ZA13 B4HDS7 B4NT24 Q7JVG2 B4PEI5 B3NCP2 A0A2M4BXR5 A0A1W4VSF0 B3M5M2 B4LEF7 B4J1R0 B4KWH1 A0A182YMT1 A0A0M4EI68 Q2LZY4 A0A182JTT7 A0A182PTD2 B4N695 A0A182W911 A0A3B0KNM4 K7J321 A0A0L7QW75 A0A0M9ADI1 A0A2A3E1L9 A0A088AVX1 A0A310SAI9 A0A154P8V3 A0A023EKD9 A0A158NFR5 B4HCT6 A0A0J7L4T4 E2A729 A0A195CJM5 A0A195B3I3 A0A195EU66 A0A023F9Z3 A0A232FE79 A0A224XMM0 A0A0V0GCF9 A0A069DW97 F4W494 A0A336LMD7 E2C865 A0A1B6DJG1
A0A0N1IH85 A0A194Q176 A0A0L7L6P7 A0A0A1XQH5 A0A2W1BMN0 W8B688 A0A0K8WHD0 A0A034VTR0 A0A0K8TR98 T1PDT3 A0A1A9W4M9 D6WDJ7 U5EVU2 A0A0L0C1T9 A0A1Y1L4X1 T1DIN2 D3TM32 T1E2Y0 A0A1L8DLT2 A0A1W4XMX2 A0A1B0G5R0 A0A1A9ZMT9 A0A146M500 A0A1Q3EU50 A0A1S4JVA9 A0A182INR5 A0A1Q3EU45 A0A182M0R3 A0A1Q3EU35 A0A182GB38 A0A182RR76 A0A1I8PP03 A0A182U3C1 A0A182NAR9 A0A182UM49 A0A2C9GR99 A0A182KTK5 Q7Q196 A0A182XJK9 A0A182Q1R7 A0A182KGK4 A0A2M4BXZ3 A0A182F3V4 W5JW34 A0A2M4ANY6 A0A2M4AN06 A0A2M4ANQ2 A0A2M3ZI64 A0A1B0CHT3 Q17EU8 A0A2M3ZA13 B4HDS7 B4NT24 Q7JVG2 B4PEI5 B3NCP2 A0A2M4BXR5 A0A1W4VSF0 B3M5M2 B4LEF7 B4J1R0 B4KWH1 A0A182YMT1 A0A0M4EI68 Q2LZY4 A0A182JTT7 A0A182PTD2 B4N695 A0A182W911 A0A3B0KNM4 K7J321 A0A0L7QW75 A0A0M9ADI1 A0A2A3E1L9 A0A088AVX1 A0A310SAI9 A0A154P8V3 A0A023EKD9 A0A158NFR5 B4HCT6 A0A0J7L4T4 E2A729 A0A195CJM5 A0A195B3I3 A0A195EU66 A0A023F9Z3 A0A232FE79 A0A224XMM0 A0A0V0GCF9 A0A069DW97 F4W494 A0A336LMD7 E2C865 A0A1B6DJG1
Pubmed
19121390
28756777
22118469
26354079
26227816
25830018
+ More
24495485 25348373 26369729 25315136 18362917 19820115 26108605 28004739 24330624 20353571 26823975 26483478 20966253 12364791 14747013 17210077 20920257 23761445 17510324 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20394834 26109357 26109356 17550304 25244985 15632085 20075255 24945155 21347285 20798317 25474469 28648823 26334808 21719571
24495485 25348373 26369729 25315136 18362917 19820115 26108605 28004739 24330624 20353571 26823975 26483478 20966253 12364791 14747013 17210077 20920257 23761445 17510324 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20394834 26109357 26109356 17550304 25244985 15632085 20075255 24945155 21347285 20798317 25474469 28648823 26334808 21719571
EMBL
BABH01028609
ODYU01005074
SOQ45596.1
KZ149897
PZC78693.1
NWSH01001020
+ More
PCG73077.1 GDQN01002220 JAT88834.1 AGBW02014545 OWR41470.1 KQ459692 KPJ20423.1 KQ459590 KPI97150.1 JTDY01002659 KOB70991.1 GBXI01001026 JAD13266.1 KZ149961 PZC76332.1 GAMC01014009 GAMC01014007 JAB92548.1 GDHF01028273 GDHF01001751 JAI24041.1 JAI50563.1 GAKP01013767 GAKP01013766 JAC45185.1 GDAI01000706 JAI16897.1 KA646906 AFP61535.1 KQ971322 EFA00788.1 GANO01001725 JAB58146.1 JRES01000997 KNC26280.1 GEZM01064843 JAV68634.1 GALA01000856 JAA93996.1 EZ422484 ADD18760.1 GALA01000880 JAA93972.1 GFDF01006666 JAV07418.1 CCAG010018755 GDHC01004979 JAQ13650.1 GFDL01016212 JAV18833.1 GFDL01016215 JAV18830.1 AXCM01008285 GFDL01016220 JAV18825.1 JXUM01157546 KQ572780 KXJ67997.1 APCN01001706 AAAB01008980 EAA14066.3 AXCN02000660 GGFJ01008670 MBW57811.1 ADMH02000223 ETN67330.1 GGFK01009007 MBW42328.1 GGFK01008843 MBW42164.1 GGFK01008917 MBW42238.1 GGFM01007506 MBW28257.1 AJWK01012667 CH477279 EAT45003.1 GGFM01004527 MBW25278.1 CH480815 EDW41017.1 CH982634 CM002912 EDX15714.1 KMY98895.1 AE014296 AY118969 AJ863561 AJ863562 AJ863563 AJ863564 AAF50123.1 AAM50829.1 AAN11888.1 AAN11889.1 CAI10728.1 CM000159 EDW94051.1 CH954178 EDV51339.1 GGFJ01008671 MBW57812.1 CH902618 EDV40656.2 CH940647 EDW70133.1 CH916366 EDV96980.1 CH933809 EDW19600.1 CP012525 ALC43501.1 CH379069 EAL31153.1 CH964154 EDW79884.1 OUUW01000009 SPP85428.1 KQ414716 KOC62852.1 KQ435692 KOX81073.1 KZ288488 PBC25222.1 KQ784160 OAD51874.1 KQ434827 KZC07570.1 GAPW01003992 JAC09606.1 ADTU01001536 CH479433 EDW30692.1 LBMM01000724 KMQ97626.1 GL437252 EFN70761.1 KQ977642 KYN00918.1 KQ976625 KYM79053.1 KQ981979 KYN31447.1 GBBI01000969 JAC17743.1 NNAY01000371 OXU28833.1 GFTR01002680 JAW13746.1 GECL01000378 JAP05746.1 GBGD01003290 JAC85599.1 GL887491 EGI71090.1 UFQS01000016 UFQT01000016 SSW97403.1 SSX17789.1 GL453568 EFN75868.1 GEDC01011506 JAS25792.1
PCG73077.1 GDQN01002220 JAT88834.1 AGBW02014545 OWR41470.1 KQ459692 KPJ20423.1 KQ459590 KPI97150.1 JTDY01002659 KOB70991.1 GBXI01001026 JAD13266.1 KZ149961 PZC76332.1 GAMC01014009 GAMC01014007 JAB92548.1 GDHF01028273 GDHF01001751 JAI24041.1 JAI50563.1 GAKP01013767 GAKP01013766 JAC45185.1 GDAI01000706 JAI16897.1 KA646906 AFP61535.1 KQ971322 EFA00788.1 GANO01001725 JAB58146.1 JRES01000997 KNC26280.1 GEZM01064843 JAV68634.1 GALA01000856 JAA93996.1 EZ422484 ADD18760.1 GALA01000880 JAA93972.1 GFDF01006666 JAV07418.1 CCAG010018755 GDHC01004979 JAQ13650.1 GFDL01016212 JAV18833.1 GFDL01016215 JAV18830.1 AXCM01008285 GFDL01016220 JAV18825.1 JXUM01157546 KQ572780 KXJ67997.1 APCN01001706 AAAB01008980 EAA14066.3 AXCN02000660 GGFJ01008670 MBW57811.1 ADMH02000223 ETN67330.1 GGFK01009007 MBW42328.1 GGFK01008843 MBW42164.1 GGFK01008917 MBW42238.1 GGFM01007506 MBW28257.1 AJWK01012667 CH477279 EAT45003.1 GGFM01004527 MBW25278.1 CH480815 EDW41017.1 CH982634 CM002912 EDX15714.1 KMY98895.1 AE014296 AY118969 AJ863561 AJ863562 AJ863563 AJ863564 AAF50123.1 AAM50829.1 AAN11888.1 AAN11889.1 CAI10728.1 CM000159 EDW94051.1 CH954178 EDV51339.1 GGFJ01008671 MBW57812.1 CH902618 EDV40656.2 CH940647 EDW70133.1 CH916366 EDV96980.1 CH933809 EDW19600.1 CP012525 ALC43501.1 CH379069 EAL31153.1 CH964154 EDW79884.1 OUUW01000009 SPP85428.1 KQ414716 KOC62852.1 KQ435692 KOX81073.1 KZ288488 PBC25222.1 KQ784160 OAD51874.1 KQ434827 KZC07570.1 GAPW01003992 JAC09606.1 ADTU01001536 CH479433 EDW30692.1 LBMM01000724 KMQ97626.1 GL437252 EFN70761.1 KQ977642 KYN00918.1 KQ976625 KYM79053.1 KQ981979 KYN31447.1 GBBI01000969 JAC17743.1 NNAY01000371 OXU28833.1 GFTR01002680 JAW13746.1 GECL01000378 JAP05746.1 GBGD01003290 JAC85599.1 GL887491 EGI71090.1 UFQS01000016 UFQT01000016 SSW97403.1 SSX17789.1 GL453568 EFN75868.1 GEDC01011506 JAS25792.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000095301 UP000091820 UP000007266 UP000037069 UP000192223 UP000092444 UP000092445 UP000075880 UP000075883 UP000069940 UP000249989 UP000075900 UP000095300 UP000075902 UP000075884 UP000075903 UP000075840 UP000075882 UP000007062 UP000076407 UP000075886 UP000075881 UP000069272 UP000000673 UP000092461 UP000008820 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000192221 UP000007801 UP000008792 UP000001070 UP000009192 UP000076408 UP000092553 UP000001819 UP000075885 UP000007798 UP000075920 UP000268350 UP000002358 UP000053825 UP000053105 UP000242457 UP000005203 UP000076502 UP000005205 UP000008744 UP000036403 UP000000311 UP000078542 UP000078540 UP000078541 UP000215335 UP000007755 UP000008237
UP000095301 UP000091820 UP000007266 UP000037069 UP000192223 UP000092444 UP000092445 UP000075880 UP000075883 UP000069940 UP000249989 UP000075900 UP000095300 UP000075902 UP000075884 UP000075903 UP000075840 UP000075882 UP000007062 UP000076407 UP000075886 UP000075881 UP000069272 UP000000673 UP000092461 UP000008820 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000192221 UP000007801 UP000008792 UP000001070 UP000009192 UP000076408 UP000092553 UP000001819 UP000075885 UP000007798 UP000075920 UP000268350 UP000002358 UP000053825 UP000053105 UP000242457 UP000005203 UP000076502 UP000005205 UP000008744 UP000036403 UP000000311 UP000078542 UP000078540 UP000078541 UP000215335 UP000007755 UP000008237
Interpro
IPR000086
NUDIX_hydrolase_dom
+ More
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR020084 NUDIX_hydrolase_CS
IPR020476 Nudix_hydrolase
IPR018957 Znf_C3HC4_RING-type
IPR013083 Znf_RING/FYVE/PHD
IPR004170 WWE-dom
IPR017907 Znf_RING_CS
IPR001841 Znf_RING
IPR018123 WWE-dom_subgr
IPR033509 RNF146
IPR037197 WWE_dom_sf
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR020084 NUDIX_hydrolase_CS
IPR020476 Nudix_hydrolase
IPR018957 Znf_C3HC4_RING-type
IPR013083 Znf_RING/FYVE/PHD
IPR004170 WWE-dom
IPR017907 Znf_RING_CS
IPR001841 Znf_RING
IPR018123 WWE-dom_subgr
IPR033509 RNF146
IPR037197 WWE_dom_sf
Gene 3D
ProteinModelPortal
H9JR13
A0A2H1VXY9
A0A2W1C3F6
A0A2A4JLZ0
A0A1E1WPC9
A0A212EJ29
+ More
A0A0N1IH85 A0A194Q176 A0A0L7L6P7 A0A0A1XQH5 A0A2W1BMN0 W8B688 A0A0K8WHD0 A0A034VTR0 A0A0K8TR98 T1PDT3 A0A1A9W4M9 D6WDJ7 U5EVU2 A0A0L0C1T9 A0A1Y1L4X1 T1DIN2 D3TM32 T1E2Y0 A0A1L8DLT2 A0A1W4XMX2 A0A1B0G5R0 A0A1A9ZMT9 A0A146M500 A0A1Q3EU50 A0A1S4JVA9 A0A182INR5 A0A1Q3EU45 A0A182M0R3 A0A1Q3EU35 A0A182GB38 A0A182RR76 A0A1I8PP03 A0A182U3C1 A0A182NAR9 A0A182UM49 A0A2C9GR99 A0A182KTK5 Q7Q196 A0A182XJK9 A0A182Q1R7 A0A182KGK4 A0A2M4BXZ3 A0A182F3V4 W5JW34 A0A2M4ANY6 A0A2M4AN06 A0A2M4ANQ2 A0A2M3ZI64 A0A1B0CHT3 Q17EU8 A0A2M3ZA13 B4HDS7 B4NT24 Q7JVG2 B4PEI5 B3NCP2 A0A2M4BXR5 A0A1W4VSF0 B3M5M2 B4LEF7 B4J1R0 B4KWH1 A0A182YMT1 A0A0M4EI68 Q2LZY4 A0A182JTT7 A0A182PTD2 B4N695 A0A182W911 A0A3B0KNM4 K7J321 A0A0L7QW75 A0A0M9ADI1 A0A2A3E1L9 A0A088AVX1 A0A310SAI9 A0A154P8V3 A0A023EKD9 A0A158NFR5 B4HCT6 A0A0J7L4T4 E2A729 A0A195CJM5 A0A195B3I3 A0A195EU66 A0A023F9Z3 A0A232FE79 A0A224XMM0 A0A0V0GCF9 A0A069DW97 F4W494 A0A336LMD7 E2C865 A0A1B6DJG1
A0A0N1IH85 A0A194Q176 A0A0L7L6P7 A0A0A1XQH5 A0A2W1BMN0 W8B688 A0A0K8WHD0 A0A034VTR0 A0A0K8TR98 T1PDT3 A0A1A9W4M9 D6WDJ7 U5EVU2 A0A0L0C1T9 A0A1Y1L4X1 T1DIN2 D3TM32 T1E2Y0 A0A1L8DLT2 A0A1W4XMX2 A0A1B0G5R0 A0A1A9ZMT9 A0A146M500 A0A1Q3EU50 A0A1S4JVA9 A0A182INR5 A0A1Q3EU45 A0A182M0R3 A0A1Q3EU35 A0A182GB38 A0A182RR76 A0A1I8PP03 A0A182U3C1 A0A182NAR9 A0A182UM49 A0A2C9GR99 A0A182KTK5 Q7Q196 A0A182XJK9 A0A182Q1R7 A0A182KGK4 A0A2M4BXZ3 A0A182F3V4 W5JW34 A0A2M4ANY6 A0A2M4AN06 A0A2M4ANQ2 A0A2M3ZI64 A0A1B0CHT3 Q17EU8 A0A2M3ZA13 B4HDS7 B4NT24 Q7JVG2 B4PEI5 B3NCP2 A0A2M4BXR5 A0A1W4VSF0 B3M5M2 B4LEF7 B4J1R0 B4KWH1 A0A182YMT1 A0A0M4EI68 Q2LZY4 A0A182JTT7 A0A182PTD2 B4N695 A0A182W911 A0A3B0KNM4 K7J321 A0A0L7QW75 A0A0M9ADI1 A0A2A3E1L9 A0A088AVX1 A0A310SAI9 A0A154P8V3 A0A023EKD9 A0A158NFR5 B4HCT6 A0A0J7L4T4 E2A729 A0A195CJM5 A0A195B3I3 A0A195EU66 A0A023F9Z3 A0A232FE79 A0A224XMM0 A0A0V0GCF9 A0A069DW97 F4W494 A0A336LMD7 E2C865 A0A1B6DJG1
PDB
5LTU
E-value=6.02253e-42,
Score=424
Ontologies
GO
GO:0016787
GO:1901911
GO:0008486
GO:0071543
GO:0000298
GO:0034432
GO:0034431
GO:1901907
GO:0050072
GO:0005634
GO:1901909
GO:0005737
GO:0009117
GO:0042594
GO:0055088
GO:0043647
GO:0042593
GO:0052842
GO:0052840
GO:0016055
GO:0061630
GO:0072572
GO:0008270
GO:0051903
GO:0016491
GO:0016310
GO:0016627
GO:0030145
GO:0015923
GO:0006260
PANTHER
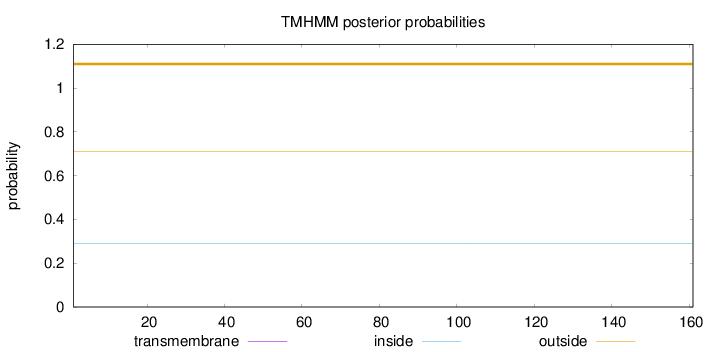
Topology
Length:
161
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.29078
outside
1 - 161
Population Genetic Test Statistics
Pi
306.465826
Theta
220.873824
Tajima's D
1.216491
CLR
0
CSRT
0.72186390680466
Interpretation
Uncertain
