Gene
KWMTBOMO06378
Pre Gene Modal
BGIBMGA011974
Annotation
PREDICTED:_uncharacterized_protein_C18orf8_[Bombyx_mori]
Full name
Tyrosine--tRNA ligase
+ More
Regulator of MON1-CCZ1 complex
Regulator of MON1-CCZ1 complex
Alternative Name
Tyrosyl-tRNA synthetase
Colon cancer-associated protein Mic1
WD repeat-containing protein 98
Colon cancer-associated protein Mic1
WD repeat-containing protein 98
Location in the cell
Mitochondrial Reliability : 0.985
Sequence
CDS
ATGGCCAGATTTAATTTGAAAGATAGTGATGAATATTACTTGACTCTCACCGAACAGCCCAAACGTTTTGACCCGGACTGCCCTGTGACAAATGTATTTTTTGACGACACTAATGGACAGGTATTCACAGTACGTTCGGGTGGAATCGGAGGCGTAACCGTAAACAGTATGGATGATAGAAAGTGTACATCTTTCCAAATGGAAGACAAAGGACCCATAATATCAATTAAATTATCACCCGACCAGAAAATATTGGCGATACAACGTAATCATGAGAGTCAGAGTGCTGCAGTAGAATTCGTGAACTTCAAGGACATGTCACCTACTAATGTAGAGTACTCTCACAGCTGTAAATGGAAGAACGCAAAAATATTAGGCTTTGTTTGGCCTAAAGCAAACGAAATTGCGTTCATAACTGATCACGGTATTGAGCTGTTTCAAGTTTTACCTGAGAAAAAACAATTGAAAAGTTTGAAGAATACCACATTTAGTGGTGCGTGGTTTTCGTGGTGTCCACAAAGCAATATAGTATTGTTAGCTGGAAACAACGGAATCCTCTTGCAGCCATTTTACCTCAACAATTCTACTATAACAAAACTACAGAAGTTAGAGTTAGATTCAATGAGACCAGTGGTAGAAAGGGACGTCTTCCTGCTCTGCTTGTGCGACACGACATGGTGCGCTGTGTTCAGGCACGGCGTATCCGCTTCTGTTGCTGCCCCCGGGCCTACCGAGGTCTGGTTGATGCCCATTACGGGCCCCAGCGGCTTCCACTACACCCACATCCTCAAGACTGGACTGGTCGGGAGATTCGCTGTCAATGTGGTCGATGACTTGGTCGTAGTGCACCACCAGTCGACACAAACCTCACAAGTGTTCGACATAATGGAAGAGTCGAAGGTAGAGAACAGTATCGCCGTTCACATCCCGCTGGTGGCAGGAGCCAGCATGCGACCGGCCCTCGTGGAGGGGCAGCCGTGTTCCATGTATTCGGGCACGTGGGTGGTGTTCCAACCCGACTACATCATAGACGCCAGGCTCGGCTGTCTGTGGCAGGTCCAGCTGCTGCCAGCGGGGCTAGCGCACTCGGTGCCAAAAGACGAGATATCGAAGATAGTGGCCGCGCTGCTCCGTCGGACGGACGGCAGGGACACCATCTACAGGATCCTCCACCAGCTGATCAGCGACGCGGGAGTCTACCTGGTGGAGCTGTCTAAGGTCTTTGACGAAATCAATTCTGTTTACAGGAAGTGGGCGGACCTGGAGGCGGCGCGGCACACAGCGGGGCAGGTGGCGCCGCGCCCCCGCGCCCCCGCCCCCGCCCGCGTGCTCGTGTCGCAGGCCGACGTGTGCGAGCAGCTGCTGCCCGCGCACTCGGACGACCGCCATCTGGTGCAGGTGGTGACGGCGTACCTGGCGTCGCTGAGCCGGCAGGAGCTGGTGGTGCAGGCGGGCACGAGCGCGGCGGGAGTACGCGCGCTGGTGGCGCGGCGCGCGGGCGGGCGGCTGCGGGCGCTGGTGCGGCGCGCGGCGCTGGCCGACTCGCGCCCGCTCGCGTGCCAGCTGCTGTCCCTGCACCGCCTGCACGCCGCCGCCGCGCAGCTCGCGCTCGACATGCTGTGGAGGCTCAAGGGACACGAGGAAGTGGTAGAAGTACTACTGAGTTGGGAGGAGCCTATCAGCGCTGTGGGGGCAGCTAAGCAGGCAGGCGGTAACGTTTGGAACAATCTACCAGCTAGGAAGCTCTTGGCAGGAGCCAAGAATGACCCTTCTGCCTTCGTGGCGGTTTACCACGCCTTGCTCAACAGAAACGACAGGCTGAGGGGATCGCCGCATTTCTTGAAAGCTGAACAATGCGACATCTACGTTGAACACTATAAGAAATTGACTTCGGAACAATGA
Protein
MARFNLKDSDEYYLTLTEQPKRFDPDCPVTNVFFDDTNGQVFTVRSGGIGGVTVNSMDDRKCTSFQMEDKGPIISIKLSPDQKILAIQRNHESQSAAVEFVNFKDMSPTNVEYSHSCKWKNAKILGFVWPKANEIAFITDHGIELFQVLPEKKQLKSLKNTTFSGAWFSWCPQSNIVLLAGNNGILLQPFYLNNSTITKLQKLELDSMRPVVERDVFLLCLCDTTWCAVFRHGVSASVAAPGPTEVWLMPITGPSGFHYTHILKTGLVGRFAVNVVDDLVVVHHQSTQTSQVFDIMEESKVENSIAVHIPLVAGASMRPALVEGQPCSMYSGTWVVFQPDYIIDARLGCLWQVQLLPAGLAHSVPKDEISKIVAALLRRTDGRDTIYRILHQLISDAGVYLVELSKVFDEINSVYRKWADLEAARHTAGQVAPRPRAPAPARVLVSQADVCEQLLPAHSDDRHLVQVVTAYLASLSRQELVVQAGTSAAGVRALVARRAGGRLRALVRRAALADSRPLACQLLSLHRLHAAAAQLALDMLWRLKGHEEVVEVLLSWEEPISAVGAAKQAGGNVWNNLPARKLLAGAKNDPSAFVAVYHALLNRNDRLRGSPHFLKAEQCDIYVEHYKKLTSEQ
Summary
Description
Componement of the CCZ1-MON1 RAB7A guanine exchange factor (GEF). Acts as a positive regulator of CCZ1-MON1A/B function necessary for endosomal/autophagic flux and efficient RAB7A localization (PubMed:29038162).
Catalytic Activity
ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + H(+) + L-tyrosyl-tRNA(Tyr)
Subunit
Found in a complex with RMC1, CCZ1 MON1A and MON1B.
Similarity
Belongs to the class-I aminoacyl-tRNA synthetase family.
Belongs to the RMC1 family.
Belongs to the RMC1 family.
Keywords
Autophagy
Complete proteome
Endosome
Lysosome
Membrane
Reference proteome
Feature
chain Regulator of MON1-CCZ1 complex
Uniprot
A0A2A4JNA9
A0A2A4JLW7
A0A2W1BYG5
A0A212EJ28
A0A2H1VXR0
A0A0N1PKG0
+ More
K7IPJ5 A0A232F423 E9INW8 A0A1L8DAT8 A0A182H294 A0A0L7R0M1 F4WH29 Q16YN5 A0A1B0CUY8 A0A154PP96 A0A067RIM8 E2BEF6 A0A195CTJ2 A0A0C9QXM8 A0A336N2B0 A0A336MM14 A0A151JWB8 A0A0L8FQH6 A0A158NT90 A0A2C9KK17 A0A195E3C8 A0A0C9RK10 E2A5A8 A0A226EYR3 A0A3L8D2M2 A0A088AP33 A0A2Y9P611 A0A1Y1MXS9 T1IWP7 A0A3B3XQN6 N6U4N5 A0A2Y9SHQ8 A0A146YIV9 A0A0L8FQN6 A0A139WKS6 V4B0J6 A0A340WLZ9 A0A087YHN7 A0A3B3V8S5 W5M0I6 A0A2U4AZ40 A0A3P8RVK3 V5IDY0 U4UBY7 A0A1W4ZTM9 A0A3B5PVT3 A0A250YKU4 A0A1S3ID23 A0A2K5Z8N1 A0A1W4WFA4 A0A3S2MGU0 A0A287A4H7 V9KK72 E1BHW7 A0A3Q1GPP9 G3QQX7 H2QEC4 F7IQV6 A0A1L8H8A2 A0A2K5RB78 F7E5Z1 Q96DM3 A0A3B4UUY0 A0A2K6DFW0 A0A2K6LAJ3 A0A0D9RYL8 A0A096N685 A0A2K6PJS9 A0A2K5MRB0 H9YZM4 H2NW17 A0A2R9AR20 A0A3Q7Q8Y6 M3VUX5 B0VZT4 G7PWI4 A0A2D4IE86 A0A195BUR0 A0A1U7Q7T4 H0XBD2 E9FQM0 A0A2K5EGB6 A0A0P5MIT7 A0A2Y9E668 A0A3B4F3H1 A0A3P9CXL8 A4IHS1 H0V7B0 A0A2U3WW73
K7IPJ5 A0A232F423 E9INW8 A0A1L8DAT8 A0A182H294 A0A0L7R0M1 F4WH29 Q16YN5 A0A1B0CUY8 A0A154PP96 A0A067RIM8 E2BEF6 A0A195CTJ2 A0A0C9QXM8 A0A336N2B0 A0A336MM14 A0A151JWB8 A0A0L8FQH6 A0A158NT90 A0A2C9KK17 A0A195E3C8 A0A0C9RK10 E2A5A8 A0A226EYR3 A0A3L8D2M2 A0A088AP33 A0A2Y9P611 A0A1Y1MXS9 T1IWP7 A0A3B3XQN6 N6U4N5 A0A2Y9SHQ8 A0A146YIV9 A0A0L8FQN6 A0A139WKS6 V4B0J6 A0A340WLZ9 A0A087YHN7 A0A3B3V8S5 W5M0I6 A0A2U4AZ40 A0A3P8RVK3 V5IDY0 U4UBY7 A0A1W4ZTM9 A0A3B5PVT3 A0A250YKU4 A0A1S3ID23 A0A2K5Z8N1 A0A1W4WFA4 A0A3S2MGU0 A0A287A4H7 V9KK72 E1BHW7 A0A3Q1GPP9 G3QQX7 H2QEC4 F7IQV6 A0A1L8H8A2 A0A2K5RB78 F7E5Z1 Q96DM3 A0A3B4UUY0 A0A2K6DFW0 A0A2K6LAJ3 A0A0D9RYL8 A0A096N685 A0A2K6PJS9 A0A2K5MRB0 H9YZM4 H2NW17 A0A2R9AR20 A0A3Q7Q8Y6 M3VUX5 B0VZT4 G7PWI4 A0A2D4IE86 A0A195BUR0 A0A1U7Q7T4 H0XBD2 E9FQM0 A0A2K5EGB6 A0A0P5MIT7 A0A2Y9E668 A0A3B4F3H1 A0A3P9CXL8 A4IHS1 H0V7B0 A0A2U3WW73
EC Number
6.1.1.1
Pubmed
28756777
22118469
26354079
20075255
28648823
21282665
+ More
26483478 21719571 17510324 24845553 20798317 21347285 15562597 30249741 28004739 23537049 18362917 19820115 23254933 25765539 23542700 28087693 30723633 24402279 19393038 22398555 16136131 27762356 17431167 14702039 16177791 15489334 17897319 23186163 29038162 25362486 25319552 22722832 17975172 22002653 21292972 25186727 21993624
26483478 21719571 17510324 24845553 20798317 21347285 15562597 30249741 28004739 23537049 18362917 19820115 23254933 25765539 23542700 28087693 30723633 24402279 19393038 22398555 16136131 27762356 17431167 14702039 16177791 15489334 17897319 23186163 29038162 25362486 25319552 22722832 17975172 22002653 21292972 25186727 21993624
EMBL
NWSH01001020
PCG73074.1
PCG73075.1
KZ149897
PZC78695.1
AGBW02014545
+ More
OWR41468.1 ODYU01005074 SOQ45598.1 KQ459692 KPJ20425.1 NNAY01001008 OXU25511.1 GL764397 EFZ17717.1 GFDF01010542 JAV03542.1 JXUM01104940 KQ564928 KXJ71556.1 KQ414670 KOC64336.1 GL888148 EGI66508.1 CH477513 EAT39738.1 AJWK01029841 KQ435007 KZC13682.1 KK852609 KDR20306.1 GL447768 EFN86001.1 KQ977329 KYN03449.1 GBYB01008539 JAG78306.1 UFQT01003365 SSX34917.1 UFQT01001291 SSX29873.1 KQ981686 KYN37947.1 KQ427521 KOF66976.1 ADTU01025732 KQ979701 KYN19581.1 GBYB01008540 JAG78307.1 GL436894 EFN71389.1 LNIX01000001 OXA62745.1 QOIP01000014 RLU14755.1 GEZM01017806 GEZM01017804 GEZM01017803 GEZM01017802 JAV90453.1 JH431627 APGK01043065 KB741011 ENN75566.1 GCES01032212 GCES01003365 JAR54111.1 KOF66977.1 KQ971322 KYB28599.1 KB199952 ESP03633.1 AYCK01020879 AYCK01020880 AHAT01004140 GANP01011152 JAB73316.1 KB631957 ERL87460.1 GFFW01000642 JAV44146.1 CM012456 RVE59085.1 AEMK02000045 DQIR01292723 DQIR01297601 HDC48201.1 JW866360 AFO98877.1 CABD030108617 AACZ04065262 NBAG03000249 PNI61442.1 CM004469 OCT92310.1 JSUE03019310 JSUE03019311 AK057192 BC002950 BC008305 AF143536 AC010853 AC026634 AQIB01129108 AHZZ02012420 JU472336 AFH29140.1 ABGA01346099 ABGA01346100 ABGA01346101 NDHI03003366 PNJ81079.1 AJFE02106629 AJFE02106630 AJFE02106631 AJFE02106632 AANG04003548 DS231815 EDS35176.1 AQIA01032729 CM001293 EHH58807.1 IACK01094361 IACK01094363 LAA82528.1 KQ976409 KYM90931.1 AAQR03029291 AAQR03029292 AAQR03029293 AAQR03029294 AAQR03029295 AAQR03029296 AAQR03029297 GL732523 EFX90343.1 GDIQ01178704 JAK73021.1 BC135661 AAI35662.1 AAKN02020644
OWR41468.1 ODYU01005074 SOQ45598.1 KQ459692 KPJ20425.1 NNAY01001008 OXU25511.1 GL764397 EFZ17717.1 GFDF01010542 JAV03542.1 JXUM01104940 KQ564928 KXJ71556.1 KQ414670 KOC64336.1 GL888148 EGI66508.1 CH477513 EAT39738.1 AJWK01029841 KQ435007 KZC13682.1 KK852609 KDR20306.1 GL447768 EFN86001.1 KQ977329 KYN03449.1 GBYB01008539 JAG78306.1 UFQT01003365 SSX34917.1 UFQT01001291 SSX29873.1 KQ981686 KYN37947.1 KQ427521 KOF66976.1 ADTU01025732 KQ979701 KYN19581.1 GBYB01008540 JAG78307.1 GL436894 EFN71389.1 LNIX01000001 OXA62745.1 QOIP01000014 RLU14755.1 GEZM01017806 GEZM01017804 GEZM01017803 GEZM01017802 JAV90453.1 JH431627 APGK01043065 KB741011 ENN75566.1 GCES01032212 GCES01003365 JAR54111.1 KOF66977.1 KQ971322 KYB28599.1 KB199952 ESP03633.1 AYCK01020879 AYCK01020880 AHAT01004140 GANP01011152 JAB73316.1 KB631957 ERL87460.1 GFFW01000642 JAV44146.1 CM012456 RVE59085.1 AEMK02000045 DQIR01292723 DQIR01297601 HDC48201.1 JW866360 AFO98877.1 CABD030108617 AACZ04065262 NBAG03000249 PNI61442.1 CM004469 OCT92310.1 JSUE03019310 JSUE03019311 AK057192 BC002950 BC008305 AF143536 AC010853 AC026634 AQIB01129108 AHZZ02012420 JU472336 AFH29140.1 ABGA01346099 ABGA01346100 ABGA01346101 NDHI03003366 PNJ81079.1 AJFE02106629 AJFE02106630 AJFE02106631 AJFE02106632 AANG04003548 DS231815 EDS35176.1 AQIA01032729 CM001293 EHH58807.1 IACK01094361 IACK01094363 LAA82528.1 KQ976409 KYM90931.1 AAQR03029291 AAQR03029292 AAQR03029293 AAQR03029294 AAQR03029295 AAQR03029296 AAQR03029297 GL732523 EFX90343.1 GDIQ01178704 JAK73021.1 BC135661 AAI35662.1 AAKN02020644
Proteomes
UP000218220
UP000007151
UP000053240
UP000002358
UP000215335
UP000069940
+ More
UP000249989 UP000053825 UP000007755 UP000008820 UP000092461 UP000076502 UP000027135 UP000008237 UP000078542 UP000078541 UP000053454 UP000005205 UP000076420 UP000078492 UP000000311 UP000198287 UP000279307 UP000005203 UP000248483 UP000261480 UP000019118 UP000248484 UP000265000 UP000007266 UP000030746 UP000265300 UP000028760 UP000261500 UP000018468 UP000245320 UP000265080 UP000030742 UP000192224 UP000002852 UP000085678 UP000233140 UP000192223 UP000008227 UP000009136 UP000257200 UP000001519 UP000002277 UP000008225 UP000186698 UP000233040 UP000006718 UP000005640 UP000261420 UP000233120 UP000233180 UP000029965 UP000028761 UP000233200 UP000233060 UP000001595 UP000240080 UP000286641 UP000011712 UP000002320 UP000009130 UP000233100 UP000078540 UP000189706 UP000005225 UP000000305 UP000233020 UP000248480 UP000261460 UP000265160 UP000005447 UP000245340
UP000249989 UP000053825 UP000007755 UP000008820 UP000092461 UP000076502 UP000027135 UP000008237 UP000078542 UP000078541 UP000053454 UP000005205 UP000076420 UP000078492 UP000000311 UP000198287 UP000279307 UP000005203 UP000248483 UP000261480 UP000019118 UP000248484 UP000265000 UP000007266 UP000030746 UP000265300 UP000028760 UP000261500 UP000018468 UP000245320 UP000265080 UP000030742 UP000192224 UP000002852 UP000085678 UP000233140 UP000192223 UP000008227 UP000009136 UP000257200 UP000001519 UP000002277 UP000008225 UP000186698 UP000233040 UP000006718 UP000005640 UP000261420 UP000233120 UP000233180 UP000029965 UP000028761 UP000233200 UP000233060 UP000001595 UP000240080 UP000286641 UP000011712 UP000002320 UP000009130 UP000233100 UP000078540 UP000189706 UP000005225 UP000000305 UP000233020 UP000248480 UP000261460 UP000265160 UP000005447 UP000245340
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
A0A2A4JNA9
A0A2A4JLW7
A0A2W1BYG5
A0A212EJ28
A0A2H1VXR0
A0A0N1PKG0
+ More
K7IPJ5 A0A232F423 E9INW8 A0A1L8DAT8 A0A182H294 A0A0L7R0M1 F4WH29 Q16YN5 A0A1B0CUY8 A0A154PP96 A0A067RIM8 E2BEF6 A0A195CTJ2 A0A0C9QXM8 A0A336N2B0 A0A336MM14 A0A151JWB8 A0A0L8FQH6 A0A158NT90 A0A2C9KK17 A0A195E3C8 A0A0C9RK10 E2A5A8 A0A226EYR3 A0A3L8D2M2 A0A088AP33 A0A2Y9P611 A0A1Y1MXS9 T1IWP7 A0A3B3XQN6 N6U4N5 A0A2Y9SHQ8 A0A146YIV9 A0A0L8FQN6 A0A139WKS6 V4B0J6 A0A340WLZ9 A0A087YHN7 A0A3B3V8S5 W5M0I6 A0A2U4AZ40 A0A3P8RVK3 V5IDY0 U4UBY7 A0A1W4ZTM9 A0A3B5PVT3 A0A250YKU4 A0A1S3ID23 A0A2K5Z8N1 A0A1W4WFA4 A0A3S2MGU0 A0A287A4H7 V9KK72 E1BHW7 A0A3Q1GPP9 G3QQX7 H2QEC4 F7IQV6 A0A1L8H8A2 A0A2K5RB78 F7E5Z1 Q96DM3 A0A3B4UUY0 A0A2K6DFW0 A0A2K6LAJ3 A0A0D9RYL8 A0A096N685 A0A2K6PJS9 A0A2K5MRB0 H9YZM4 H2NW17 A0A2R9AR20 A0A3Q7Q8Y6 M3VUX5 B0VZT4 G7PWI4 A0A2D4IE86 A0A195BUR0 A0A1U7Q7T4 H0XBD2 E9FQM0 A0A2K5EGB6 A0A0P5MIT7 A0A2Y9E668 A0A3B4F3H1 A0A3P9CXL8 A4IHS1 H0V7B0 A0A2U3WW73
K7IPJ5 A0A232F423 E9INW8 A0A1L8DAT8 A0A182H294 A0A0L7R0M1 F4WH29 Q16YN5 A0A1B0CUY8 A0A154PP96 A0A067RIM8 E2BEF6 A0A195CTJ2 A0A0C9QXM8 A0A336N2B0 A0A336MM14 A0A151JWB8 A0A0L8FQH6 A0A158NT90 A0A2C9KK17 A0A195E3C8 A0A0C9RK10 E2A5A8 A0A226EYR3 A0A3L8D2M2 A0A088AP33 A0A2Y9P611 A0A1Y1MXS9 T1IWP7 A0A3B3XQN6 N6U4N5 A0A2Y9SHQ8 A0A146YIV9 A0A0L8FQN6 A0A139WKS6 V4B0J6 A0A340WLZ9 A0A087YHN7 A0A3B3V8S5 W5M0I6 A0A2U4AZ40 A0A3P8RVK3 V5IDY0 U4UBY7 A0A1W4ZTM9 A0A3B5PVT3 A0A250YKU4 A0A1S3ID23 A0A2K5Z8N1 A0A1W4WFA4 A0A3S2MGU0 A0A287A4H7 V9KK72 E1BHW7 A0A3Q1GPP9 G3QQX7 H2QEC4 F7IQV6 A0A1L8H8A2 A0A2K5RB78 F7E5Z1 Q96DM3 A0A3B4UUY0 A0A2K6DFW0 A0A2K6LAJ3 A0A0D9RYL8 A0A096N685 A0A2K6PJS9 A0A2K5MRB0 H9YZM4 H2NW17 A0A2R9AR20 A0A3Q7Q8Y6 M3VUX5 B0VZT4 G7PWI4 A0A2D4IE86 A0A195BUR0 A0A1U7Q7T4 H0XBD2 E9FQM0 A0A2K5EGB6 A0A0P5MIT7 A0A2Y9E668 A0A3B4F3H1 A0A3P9CXL8 A4IHS1 H0V7B0 A0A2U3WW73
Ontologies
GO
PANTHER
Topology
Subcellular location
Lysosome membrane
Late endosome membrane
Late endosome membrane
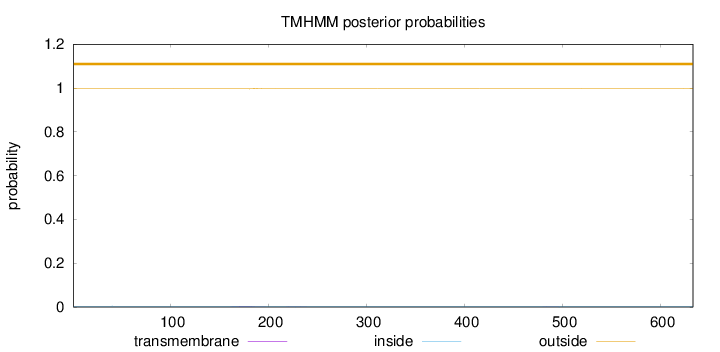
Length:
633
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07425
Exp number, first 60 AAs:
0.00105
Total prob of N-in:
0.00378
outside
1 - 633
Population Genetic Test Statistics
Pi
185.070343
Theta
154.954151
Tajima's D
0.94714
CLR
0.701324
CSRT
0.645017749112544
Interpretation
Uncertain
