Gene
KWMTBOMO06368
Pre Gene Modal
BGIBMGA011918
Annotation
PREDICTED:_D(3)_dopamine_receptor-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.982
Sequence
CDS
ATGGACGGTGCATTCGAGAACGAAAGCGACCGGGGATGGAATTCGACCGAAGAGGGGACGGATGCGAACTTGTACAATTACTGGGCTTGGAGCATCGTCGACGGTTCCCTCATGGTGTTGATCGTGAGCGGGAACACTCTCACCATCCTCGCCGTCACGCTAAGCAGAAAATTATCATGTTTAGTATCCAACCAGTTCGTACTAAACCTGGCGATATCAGATCTAATGGTCGGCCTCACCCTGCCGTACCACCTCGTTTTCTACCTGGACGATGAATTCGGAACGGTCAAATGGACGTGCTTAATGAGATTCATTCTCATAATATTAGCATGTTTGGCGTCCATTTACAACATCATAGCTATAGCCGTAGACAGGTACATAGCGATAGTGCACCCGCTCCATTACAGCCGGTACATGACAAAGCTGGTGACTCGAGTGCTGATGAGCGCCACGTGGTCAGTGGCCGTGTGCATCAGCTGCATACCGATATTCTGGAACGAGTGGCATGACGGAATTGCATGCGAAATGAACATGGTAGTACCGAAGTCATACACGACGAGCATCTTAGCGCCAATGTTCTCTTTGATATGGATGGTCATGTTCATCCTTTACTGGCGGATATGGCGCGAGGCAACTTGCCACGCAAGACGGATGCGAGCCAACACTTGTTGCCCTACTGGAGCCAATGACTGGAAGAGTATACAGGTCGTACTGCTGGTTCTTGGATCGTTTTCCATATGCTGGATGCCGTTCGTGGTAGTGGCTTGCGCACAGACCTTGCCAATTATGCCGTTACACAATCACATCGCTTACAGACTAACCTCTTCACTGGCCATGTCCAATTCAGGAATCAATCCTCTTATTTACGCTTGGAAAAATGCTGGATTCCGTGCGGCTTTCGCTAAACTGCTACGTTGCAAGCGACCTGACACATCGGAATACAGAGGATCACCGGCACCGGAAAGGAAGAGAGGATCCGTGGCTTTACGTGAAGGCTCTATCACTCGATCTAGCGCTCCGGCCGCTCTATCCAGCATGGGTGGTCGTCCTGCAAGACTGTTGTATGTCGAACAGGATTCAGATACATCTAGATGCCGCATCATTGAAAACGCTGGGTATGTTGATTCGGAGAGGGGTGATTCCAACCCGTCTTATGCTCCCGACGGTCCGACACCCGTCCACAATAAGACCGCGGCCTGCTCCCCGGACGCAGTGTAG
Protein
MDGAFENESDRGWNSTEEGTDANLYNYWAWSIVDGSLMVLIVSGNTLTILAVTLSRKLSCLVSNQFVLNLAISDLMVGLTLPYHLVFYLDDEFGTVKWTCLMRFILIILACLASIYNIIAIAVDRYIAIVHPLHYSRYMTKLVTRVLMSATWSVAVCISCIPIFWNEWHDGIACEMNMVVPKSYTTSILAPMFSLIWMVMFILYWRIWREATCHARRMRANTCCPTGANDWKSIQVVLLVLGSFSICWMPFVVVACAQTLPIMPLHNHIAYRLTSSLAMSNSGINPLIYAWKNAGFRAAFAKLLRCKRPDTSEYRGSPAPERKRGSVALREGSITRSSAPAALSSMGGRPARLLYVEQDSDTSRCRIIENAGYVDSERGDSNPSYAPDGPTPVHNKTAACSPDAV
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
H9JQW2
A0A2W1BYH6
A0A2H1WQX4
A0A3S2LZ50
A0A194PUY8
A0A0N1ID35
+ More
A0A212EJ01 A0A0L7L200 A0A1J1HRR9 B0W7U2 A0A1B0GHV7 Q16RJ5 U5N4G2 A0A1L8DJS7 A0A2J7QAS8 A0A182GK28 A0A336LTV7 A0A067R1B0 A0A2P8YQF6 A0A151WSG2 A0A151I582 A0A1Y1MBS0 A0A0L0BX80 A0A182RHP4 A0A182IPG4 A0A182W5R1 A0A182PT70 A0A1I8MPD5 A0A151JAI8 A0A195FL79 A0A0J7NZ83 N6TLI0 A0A182N0J0 A0A195C844 A0A232F583 B4N1Y2 A0A182T4Z6 D6WF87 N1NVF1 A0A109NH66 B3MQW1 A0A1W4V830 Q9W3V5 B4R5Q2 A0A3B0K5W3 B4I044 B4PZ89 B4GWJ1 B4M6Z2 B3NXW7 Q29I79 A0A1W4WMV9 A0A1B6MIK0 A0A026X2D4 A0A0M5J161 B4JXH9 A0A0K8U5D5 A0A1A9WAZ1 E0VAP6 A0A1S3JRB9 A0A1S3H9Q3 A0A3Q2YVD4 A0A1S3HRK7 K1PWL3 A0A1S3IU64 A0A3P8T270 A0A210R506 A0A3Q2XUJ8 A0A3Q0R5Q2 A0A1L8GXX4 A0A3B3ZSG4 A0A1L8GV34 A0A0L8GUP3 F7ECL3 A0A1S3JUE9 A0A3B5AUM8
A0A212EJ01 A0A0L7L200 A0A1J1HRR9 B0W7U2 A0A1B0GHV7 Q16RJ5 U5N4G2 A0A1L8DJS7 A0A2J7QAS8 A0A182GK28 A0A336LTV7 A0A067R1B0 A0A2P8YQF6 A0A151WSG2 A0A151I582 A0A1Y1MBS0 A0A0L0BX80 A0A182RHP4 A0A182IPG4 A0A182W5R1 A0A182PT70 A0A1I8MPD5 A0A151JAI8 A0A195FL79 A0A0J7NZ83 N6TLI0 A0A182N0J0 A0A195C844 A0A232F583 B4N1Y2 A0A182T4Z6 D6WF87 N1NVF1 A0A109NH66 B3MQW1 A0A1W4V830 Q9W3V5 B4R5Q2 A0A3B0K5W3 B4I044 B4PZ89 B4GWJ1 B4M6Z2 B3NXW7 Q29I79 A0A1W4WMV9 A0A1B6MIK0 A0A026X2D4 A0A0M5J161 B4JXH9 A0A0K8U5D5 A0A1A9WAZ1 E0VAP6 A0A1S3JRB9 A0A1S3H9Q3 A0A3Q2YVD4 A0A1S3HRK7 K1PWL3 A0A1S3IU64 A0A3P8T270 A0A210R506 A0A3Q2XUJ8 A0A3Q0R5Q2 A0A1L8GXX4 A0A3B3ZSG4 A0A1L8GV34 A0A0L8GUP3 F7ECL3 A0A1S3JUE9 A0A3B5AUM8
Pubmed
19121390
28756777
26354079
22118469
26227816
17510324
+ More
24130914 26483478 24845553 29403074 28004739 26108605 25315136 23537049 28648823 17994087 18362917 19820115 18054377 18025266 18316733 20068045 21843505 23604020 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 24508170 20566863 22992520 28812685 27762356 25463417 20431018
24130914 26483478 24845553 29403074 28004739 26108605 25315136 23537049 28648823 17994087 18362917 19820115 18054377 18025266 18316733 20068045 21843505 23604020 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 24508170 20566863 22992520 28812685 27762356 25463417 20431018
EMBL
BABH01028611
KZ149897
PZC78705.1
ODYU01010366
SOQ55428.1
RSAL01000104
+ More
RVE47355.1 KQ459590 KPI97137.1 KQ459692 KPJ20435.1 AGBW02014545 OWR41459.1 JTDY01003478 KOB69497.1 CVRI01000020 CRK90717.1 DS231855 EDS38236.1 AJWK01005694 CH477708 EAT37025.1 KC439532 AGX85000.1 GFDF01007490 JAV06594.1 NEVH01016327 PNF25682.1 JXUM01013072 KQ560368 KXJ82762.1 UFQT01000107 SSX20069.1 KK852999 KDR12658.1 PYGN01000433 PSN46479.1 KQ982775 KYQ50768.1 KQ976445 KYM86129.1 GEZM01039614 JAV81316.1 JRES01001295 KNC23839.1 KQ979275 KYN22037.1 KQ981522 KYN40744.1 LBMM01000708 KMQ97680.1 APGK01019725 KB740120 KB631607 ENN81314.1 ERL84634.1 KQ978143 KYM96820.1 NNAY01000979 OXU25638.1 CH963925 EDW78371.1 KQ971319 EFA01319.1 BK005870 DAA64507.1 JQ673411 AFJ19499.1 CH902622 EDV34166.1 AE014298 BT011376 AAF46208.1 AAR96168.1 CM000366 EDX17289.1 OUUW01000003 SPP78848.1 CH480819 EDW52875.1 CM000162 EDX01056.1 CH479194 EDW27075.1 CH940653 EDW62559.1 CH954180 EDV47418.1 CH379063 EAL32774.3 GEBQ01004255 JAT35722.1 KK107020 EZA62475.1 CP012528 ALC49236.1 CH916376 EDV95455.1 GDHF01030724 JAI21590.1 DS235012 EEB10452.1 JH818700 EKC23414.1 NEDP02000279 OWF56153.1 CM004470 OCT88651.1 CM004471 OCT87695.1 KQ420273 KOF80776.1 AAMC01076508
RVE47355.1 KQ459590 KPI97137.1 KQ459692 KPJ20435.1 AGBW02014545 OWR41459.1 JTDY01003478 KOB69497.1 CVRI01000020 CRK90717.1 DS231855 EDS38236.1 AJWK01005694 CH477708 EAT37025.1 KC439532 AGX85000.1 GFDF01007490 JAV06594.1 NEVH01016327 PNF25682.1 JXUM01013072 KQ560368 KXJ82762.1 UFQT01000107 SSX20069.1 KK852999 KDR12658.1 PYGN01000433 PSN46479.1 KQ982775 KYQ50768.1 KQ976445 KYM86129.1 GEZM01039614 JAV81316.1 JRES01001295 KNC23839.1 KQ979275 KYN22037.1 KQ981522 KYN40744.1 LBMM01000708 KMQ97680.1 APGK01019725 KB740120 KB631607 ENN81314.1 ERL84634.1 KQ978143 KYM96820.1 NNAY01000979 OXU25638.1 CH963925 EDW78371.1 KQ971319 EFA01319.1 BK005870 DAA64507.1 JQ673411 AFJ19499.1 CH902622 EDV34166.1 AE014298 BT011376 AAF46208.1 AAR96168.1 CM000366 EDX17289.1 OUUW01000003 SPP78848.1 CH480819 EDW52875.1 CM000162 EDX01056.1 CH479194 EDW27075.1 CH940653 EDW62559.1 CH954180 EDV47418.1 CH379063 EAL32774.3 GEBQ01004255 JAT35722.1 KK107020 EZA62475.1 CP012528 ALC49236.1 CH916376 EDV95455.1 GDHF01030724 JAI21590.1 DS235012 EEB10452.1 JH818700 EKC23414.1 NEDP02000279 OWF56153.1 CM004470 OCT88651.1 CM004471 OCT87695.1 KQ420273 KOF80776.1 AAMC01076508
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000183832 UP000002320 UP000092461 UP000008820 UP000235965 UP000069940 UP000249989 UP000027135 UP000245037 UP000075809 UP000078540 UP000037069 UP000075900 UP000075880 UP000075920 UP000075885 UP000095301 UP000078492 UP000078541 UP000036403 UP000019118 UP000030742 UP000075884 UP000078542 UP000215335 UP000007798 UP000075901 UP000007266 UP000007801 UP000192221 UP000000803 UP000000304 UP000268350 UP000001292 UP000002282 UP000008744 UP000008792 UP000008711 UP000001819 UP000192223 UP000053097 UP000092553 UP000001070 UP000091820 UP000009046 UP000085678 UP000264820 UP000005408 UP000265080 UP000242188 UP000261340 UP000186698 UP000261520 UP000053454 UP000008143 UP000261400
UP000183832 UP000002320 UP000092461 UP000008820 UP000235965 UP000069940 UP000249989 UP000027135 UP000245037 UP000075809 UP000078540 UP000037069 UP000075900 UP000075880 UP000075920 UP000075885 UP000095301 UP000078492 UP000078541 UP000036403 UP000019118 UP000030742 UP000075884 UP000078542 UP000215335 UP000007798 UP000075901 UP000007266 UP000007801 UP000192221 UP000000803 UP000000304 UP000268350 UP000001292 UP000002282 UP000008744 UP000008792 UP000008711 UP000001819 UP000192223 UP000053097 UP000092553 UP000001070 UP000091820 UP000009046 UP000085678 UP000264820 UP000005408 UP000265080 UP000242188 UP000261340 UP000186698 UP000261520 UP000053454 UP000008143 UP000261400
Pfam
PF00001 7tm_1
Interpro
ProteinModelPortal
H9JQW2
A0A2W1BYH6
A0A2H1WQX4
A0A3S2LZ50
A0A194PUY8
A0A0N1ID35
+ More
A0A212EJ01 A0A0L7L200 A0A1J1HRR9 B0W7U2 A0A1B0GHV7 Q16RJ5 U5N4G2 A0A1L8DJS7 A0A2J7QAS8 A0A182GK28 A0A336LTV7 A0A067R1B0 A0A2P8YQF6 A0A151WSG2 A0A151I582 A0A1Y1MBS0 A0A0L0BX80 A0A182RHP4 A0A182IPG4 A0A182W5R1 A0A182PT70 A0A1I8MPD5 A0A151JAI8 A0A195FL79 A0A0J7NZ83 N6TLI0 A0A182N0J0 A0A195C844 A0A232F583 B4N1Y2 A0A182T4Z6 D6WF87 N1NVF1 A0A109NH66 B3MQW1 A0A1W4V830 Q9W3V5 B4R5Q2 A0A3B0K5W3 B4I044 B4PZ89 B4GWJ1 B4M6Z2 B3NXW7 Q29I79 A0A1W4WMV9 A0A1B6MIK0 A0A026X2D4 A0A0M5J161 B4JXH9 A0A0K8U5D5 A0A1A9WAZ1 E0VAP6 A0A1S3JRB9 A0A1S3H9Q3 A0A3Q2YVD4 A0A1S3HRK7 K1PWL3 A0A1S3IU64 A0A3P8T270 A0A210R506 A0A3Q2XUJ8 A0A3Q0R5Q2 A0A1L8GXX4 A0A3B3ZSG4 A0A1L8GV34 A0A0L8GUP3 F7ECL3 A0A1S3JUE9 A0A3B5AUM8
A0A212EJ01 A0A0L7L200 A0A1J1HRR9 B0W7U2 A0A1B0GHV7 Q16RJ5 U5N4G2 A0A1L8DJS7 A0A2J7QAS8 A0A182GK28 A0A336LTV7 A0A067R1B0 A0A2P8YQF6 A0A151WSG2 A0A151I582 A0A1Y1MBS0 A0A0L0BX80 A0A182RHP4 A0A182IPG4 A0A182W5R1 A0A182PT70 A0A1I8MPD5 A0A151JAI8 A0A195FL79 A0A0J7NZ83 N6TLI0 A0A182N0J0 A0A195C844 A0A232F583 B4N1Y2 A0A182T4Z6 D6WF87 N1NVF1 A0A109NH66 B3MQW1 A0A1W4V830 Q9W3V5 B4R5Q2 A0A3B0K5W3 B4I044 B4PZ89 B4GWJ1 B4M6Z2 B3NXW7 Q29I79 A0A1W4WMV9 A0A1B6MIK0 A0A026X2D4 A0A0M5J161 B4JXH9 A0A0K8U5D5 A0A1A9WAZ1 E0VAP6 A0A1S3JRB9 A0A1S3H9Q3 A0A3Q2YVD4 A0A1S3HRK7 K1PWL3 A0A1S3IU64 A0A3P8T270 A0A210R506 A0A3Q2XUJ8 A0A3Q0R5Q2 A0A1L8GXX4 A0A3B3ZSG4 A0A1L8GV34 A0A0L8GUP3 F7ECL3 A0A1S3JUE9 A0A3B5AUM8
PDB
6IBL
E-value=1.27535e-27,
Score=306
Ontologies
GO
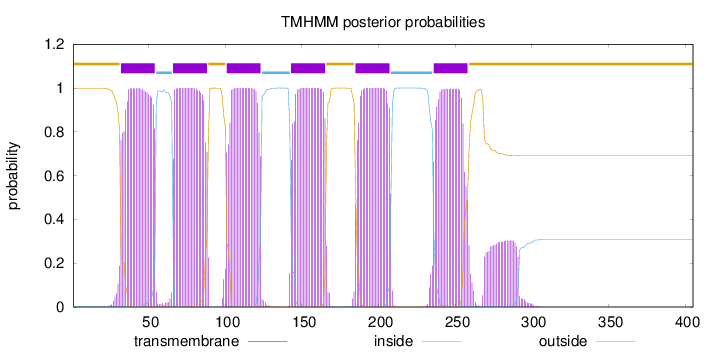
Topology
Length:
405
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
140.81382
Exp number, first 60 AAs:
22.34777
Total prob of N-in:
0.00139
POSSIBLE N-term signal
sequence
outside
1 - 31
TMhelix
32 - 54
inside
55 - 65
TMhelix
66 - 88
outside
89 - 100
TMhelix
101 - 123
inside
124 - 142
TMhelix
143 - 165
outside
166 - 184
TMhelix
185 - 207
inside
208 - 235
TMhelix
236 - 258
outside
259 - 405
Population Genetic Test Statistics
Pi
234.097207
Theta
206.889265
Tajima's D
0.683057
CLR
0.328105
CSRT
0.567421628918554
Interpretation
Uncertain
