Gene
KWMTBOMO06367
Pre Gene Modal
BGIBMGA011979
Annotation
PREDICTED:_serine/threonine-protein_kinase_SMG1-like_[Bombyx_mori]
Full name
Non-specific serine/threonine protein kinase
+ More
Serine/threonine-protein kinase SMG1
Serine/threonine-protein kinase SMG1
Alternative Name
61E3.4
Lambda/iota protein kinase C-interacting protein
Lambda/iota protein kinase C-interacting protein
Location in the cell
Cytoplasmic Reliability : 1.697 Golgi Reliability : 1.631
Sequence
CDS
ATGGTCCACACTGGCATATTCCGTCTCGCCTGCGAGCACGTGTTGCGCACGATGCGTCGTGGCCGCGAAACCCTGTTGACGTTGTTGGAAGCGTTCGTGTACGATCCGCTGGTCGAATGGGGCGGCAGCAGCGGAGGGAAGCGACGCCGCACGCAGAGAGACGTTAAATCGGCTTTGGACATGATGGCGGTGAGAGCACAAGAATTGCAACATAGTCTGGCCCAAGTCACAGAACAATTTCTGGCCATCTTGCCTGGTATCATTGAAAGCGCTGACAAGTGGCAAAAAGAACATGAAGAACTGGTTGAAGTCGAGGCGAGACTCCAAGATTGCCACCAACAAATGGCACTAATCAAAGAAATAGAAGCATATGGACCTAATCTGAACAACCATCCATTGCACGCAATCTCGCAAAAGTACTCTTCATACAAACAAGCTAAGAATGCTGTTGAAGATTCCAAAAAAGCGTTAGTCAAAATACTGAATGATTTTGACGCACAGATCGAAAGTTTCTCAGCGACTTCTGAACTTCTTAATGGTCCGCAATTGATGGCTTGGGTGCAAGAATTTTCCGCTCCAAACGAAGAAGAAGACACGCCGATCTTTGAACACATAAAAGATTTTCTAACAAATGCAGGTCAAAGCTCCATGATCACTCAATGCGAACAAGCCGAAAAAGAATTTTATCAATCTTTGAAACAGACCCAGTGCATCATTAGGGCTTGTCTGGAACTTTTGTCACAATACGTAGCTGTCTCTCAATATTTCCCACAGAGTCAAACGGAATACCATCGCATAGTTATGTTCCGGAAGTTCCTTGCTGCTGCTCTAGACAGCAAATCGCCCGAGGTTTGCCGAGAGGTTGCCAGTCAAGTCAACGCCATCATCAATGCGGAGAACAACAAAGGCGATCCGCAACAAATTATTGCTTATAATTATCGCTTGGAAACCATCAGTGCTAAAGCGAACGCAAACTTGGCTAAGTGTGTTGAGAAGTTGCAACTAGAAGGTGGACCAGAAGCCATGGCTGTTGCTCAAGAAGCTTACAGAGAGGCAAAGGCGAGCATCGGCAACTGGGTCAGGTCTGAAGAAGGAGCCGCAACCGCTTTAGAATGTGCTGTTATCAGCATGCTGTGTCATTTAAATCGAAGATATCTGATGCTTGAGAGCGGTGCCCAAAGTGCTGGTGATTGTCTCGTGGACTTAACGTCGAGAGAAGGAGAGTGGTTCCTAGATGATATGAGCGCTTTGTCGACCCAAGCTGTGGAGCTTTTATCGCTGCTGCCGTTGCAGTCTGCTTCAGTAGAAGACGCTGCTTTACCAGTAGCAGTGGAATGTGTTCGGAATGTGAATTACCTGCTAGCTGATTTGGTCCAACTAAATTACAATTTCAGCACGATCATACTTCCTGAAGCGTTAAAGAAGATACATTCTGAAGAACCGTCTGTCCTACTAATGATAAATGAACTGAATGTGGTCATAATGAATTCTACTGTGCCTTTGAACGAGTTATTGGCCCAGCTTGAAGTGCATCTTAGATACTTGGTCATGGATATGGAGTCTCCAGCAAGCAGCGCTCAAGTTGTAGCGGCAGAACTGCGTGCTCGTTATGAAGCACTGCTTTCTGCCCCGACTTCTGATGTGGAGGGACAGTCTTCAGGACGAATGCTGCTTATGGGTTTCAACGGTCTATTCGCTGCTGTAGAATTGAGAGGAAGGGAACTGGCCGACCACTTGGCAATACCTATACCTCCAGCTTGGAGGAAGATTGATCATATCAGCGAATCTATGCACATGTCTGTAAGTGTAGTTTGTTTGTTTGTTTATTTATATTTCTTGGCCATCAATGTTAAATAA
Protein
MVHTGIFRLACEHVLRTMRRGRETLLTLLEAFVYDPLVEWGGSSGGKRRRTQRDVKSALDMMAVRAQELQHSLAQVTEQFLAILPGIIESADKWQKEHEELVEVEARLQDCHQQMALIKEIEAYGPNLNNHPLHAISQKYSSYKQAKNAVEDSKKALVKILNDFDAQIESFSATSELLNGPQLMAWVQEFSAPNEEEDTPIFEHIKDFLTNAGQSSMITQCEQAEKEFYQSLKQTQCIIRACLELLSQYVAVSQYFPQSQTEYHRIVMFRKFLAAALDSKSPEVCREVASQVNAIINAENNKGDPQQIIAYNYRLETISAKANANLAKCVEKLQLEGGPEAMAVAQEAYREAKASIGNWVRSEEGAATALECAVISMLCHLNRRYLMLESGAQSAGDCLVDLTSREGEWFLDDMSALSTQAVELLSLLPLQSASVEDAALPVAVECVRNVNYLLADLVQLNYNFSTIILPEALKKIHSEEPSVLLMINELNVVIMNSTVPLNELLAQLEVHLRYLVMDMESPASSAQVVAAELRARYEALLSAPTSDVEGQSSGRMLLMGFNGLFAAVELRGRELADHLAIPIPPAWRKIDHISESMHMSVSVVCLFVYLYFLAINVK
Summary
Description
Serine/threonine protein kinase involved in both mRNA surveillance and genotoxic stress response pathways. Recognizes the substrate consensus sequence [ST]-Q. Plays a central role in nonsense-mediated decay (NMD) of mRNAs containing premature stop codons by phosphorylating UPF1/RENT1. Recruited by release factors to stalled ribosomes together with SMG8 and SMG9 (forming the SMG1C protein kinase complex), and UPF1 to form the transient SURF (SMG1-UPF1-eRF1-eRF3) complex. In EJC-dependent NMD, the SURF complex associates with the exon junction complex (EJC) through UPF2 and allows the formation of an UPF1-UPF2-UPF3 surveillance complex which is believed to activate NMD. Also acts as a genotoxic stress-activated protein kinase that displays some functional overlap with ATM. Can phosphorylate p53/TP53 and is required for optimal p53/TP53 activation after cellular exposure to genotoxic stress. Its depletion leads to spontaneous DNA damage and increased sensitivity to ionizing radiation (IR). May activate PRKCI but not PRKCZ (By similarity).
Serine/threonine protein kinase involved in both mRNA surveillance and genotoxic stress response pathways. Recognizes the substrate consensus sequence [ST]-Q. Plays a central role in nonsense-mediated decay (NMD) of mRNAs containing premature stop codons by phosphorylating UPF1/RENT1. Recruited by release factors to stalled ribosomes together with SMG8 and SMG9 (forming the SMG1C protein kinase complex), and UPF1 to form the transient SURF (SMG1-UPF1-eRF1-eRF3) complex. In EJC-dependent NMD, the SURF complex associates with the exon junction complex (EJC) through UPF2 and allows the formation of an UPF1-UPF2-UPF3 surveillance complex which is believed to activate NMD. Also acts as a genotoxic stress-activated protein kinase that displays some functional overlap with ATM. Can phosphorylate p53/TP53 and is required for optimal p53/TP53 activation after cellular exposure to genotoxic stress. Its depletion leads to spontaneous DNA damage and increased sensitivity to ionizing radiation (IR). May activate PRKCI but not PRKCZ.
Serine/threonine protein kinase involved in both mRNA surveillance and genotoxic stress response pathways. Recognizes the substrate consensus sequence [ST]-Q. Plays a central role in nonsense-mediated decay (NMD) of mRNAs containing premature stop codons by phosphorylating UPF1/RENT1. Recruited by release factors to stalled ribosomes together with SMG8 and SMG9 (forming the SMG1C protein kinase complex), and UPF1 to form the transient SURF (SMG1-UPF1-eRF1-eRF3) complex. In EJC-dependent NMD, the SURF complex associates with the exon junction complex (EJC) through UPF2 and allows the formation of an UPF1-UPF2-UPF3 surveillance complex which is believed to activate NMD. Also acts as a genotoxic stress-activated protein kinase that displays some functional overlap with ATM. Can phosphorylate p53/TP53 and is required for optimal p53/TP53 activation after cellular exposure to genotoxic stress. Its depletion leads to spontaneous DNA damage and increased sensitivity to ionizing radiation (IR). May activate PRKCI but not PRKCZ.
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mn(2+)
Subunit
Component of the SMG1C complex composed of SMG1, SMG8 and SMG9; the recruitment of SMG8 to SMG1 N-terminus induces a large conformational change in the SMG1 C-terminal head domain containing the catalytic domain. Component of the transient SURF (SMG1-UPF1-eRF1-eRF3) complex. Interacts with PRKCI. Interacts with TELO2 and TTI1. Interacts with RUVBL1 and RUVBL2.
Component of the SMG1C complex composed of SMG1, SMG8 and SMG9; the recruitment of SMG8 to SMG1 N-terminus induces a large conformational change in the SMG1 C-terminal head domain containing the catalytic domain. Component of the transient SURF (SMG1-UPF1-eRF1-eRF3) complex. Interacts with PRKCI. Interacts with TELO2 and TTI1. Interacts with RUVBL1 and RUVBL2. Interacts with UPF2.
Component of the SMG1C complex composed of SMG1, SMG8 and SMG9; the recruitment of SMG8 to SMG1 N-terminus induces a large conformational change in the SMG1 C-terminal head domain containing the catalytic domain. Component of the transient SURF (SMG1-UPF1-eRF1-eRF3) complex. Interacts with PRKCI. Interacts with TELO2 and TTI1. Interacts with RUVBL1 and RUVBL2. Interacts with UPF2.
Miscellaneous
This gene is located in a region of chromosome 16 that contains 2 segmental duplications. Other genes that are highly related to this exist, but they probably represent pseudogenes.
Similarity
Belongs to the PI3/PI4-kinase family.
Keywords
Acetylation
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Direct protein sequencing
DNA damage
DNA repair
Kinase
Manganese
Metal-binding
Nonsense-mediated mRNA decay
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Transferase
Polymorphism
Feature
chain Serine/threonine-protein kinase SMG1
splice variant In isoform 2.
sequence variant In dbSNP:rs12051350.
splice variant In isoform 2.
sequence variant In dbSNP:rs12051350.
Uniprot
H9JR19
A0A2A4JCX2
A0A2A4JC06
A0A2W1C160
A0A0N1PK13
A0A0L7LLT8
+ More
A0A194Q159 A0A2H1VWC4 A0A3S2NCA3 A0A212EJ17 A0A310SKL6 A0A0M9A9D8 A0A067R3Y6 A0A154P395 A0A0L7RD16 A0A087ZS52 A0A2A3EQ41 A0A0J7L5B8 A0A026X4W6 E2C545 A0A2P8YQE7 A0A0C9RCL1 E1ZVC1 A0A3L8DTG4 A0A232F5L9 K7J769 E9IH54 A0A195C7G7 A0A151JAE5 A0A151WSJ2 A0A195FK18 A0A151I5P1 A0A158NTX1 F4WU96 E0VAP7 A0A139WLQ5 A0A0T6B2K1 A0A1B6CNV0 A0A1B6CAC4 A0A1Y1N066 N6UHU4 U4U3T0 T1J1T4 T1I2T7 A0A224XCD0 J9KB53 A0A2H8TIB2 A0A2S2NXY1 A0A1L8EXP0 A0A1L8EQ71 F1PBU5 A0A093I1J3 G3TL75 A0A3Q7Y545 A0A3Q7XMV0 G1TC15 I3MMW0 A0A287CYX9 G1R0C7 A0A384CK10 A0A3Q7XEQ5 M3YPN5 W5PMR8 Q8BKX6 A0A2Y9KTD8 A0A2Y9KFV9 A0A3Q7TUJ0 A0A3Q7SEF6 J9NTC0 A0A2Y9KF75 A0A3Q7ST86 A0A2K5Q9B5 A0A2Y9HQ56 R0JWW4 F6S837 A0A3Q7MW16 A0A0B8RZ42 A0A287BD11 A0A2U3ZQP5 F1MBL6 A0A2K5EVM8 G7NPR4 A0A1U7T5V5 A0A2K5EVM1 A0A2K5EVL0 Q96Q15-2 A0A2U3WMW2 U3IWI0 H0VLT8 A0A2K5Q9U0 K9J0V7 F7F0R6 W5PMR0 A0A287B556 A0A096NRG0 A0A2J8Q5H4 A0A2K5Q9U5 A0A2R9BHL9 Q96Q15-4 G3R3L1
A0A194Q159 A0A2H1VWC4 A0A3S2NCA3 A0A212EJ17 A0A310SKL6 A0A0M9A9D8 A0A067R3Y6 A0A154P395 A0A0L7RD16 A0A087ZS52 A0A2A3EQ41 A0A0J7L5B8 A0A026X4W6 E2C545 A0A2P8YQE7 A0A0C9RCL1 E1ZVC1 A0A3L8DTG4 A0A232F5L9 K7J769 E9IH54 A0A195C7G7 A0A151JAE5 A0A151WSJ2 A0A195FK18 A0A151I5P1 A0A158NTX1 F4WU96 E0VAP7 A0A139WLQ5 A0A0T6B2K1 A0A1B6CNV0 A0A1B6CAC4 A0A1Y1N066 N6UHU4 U4U3T0 T1J1T4 T1I2T7 A0A224XCD0 J9KB53 A0A2H8TIB2 A0A2S2NXY1 A0A1L8EXP0 A0A1L8EQ71 F1PBU5 A0A093I1J3 G3TL75 A0A3Q7Y545 A0A3Q7XMV0 G1TC15 I3MMW0 A0A287CYX9 G1R0C7 A0A384CK10 A0A3Q7XEQ5 M3YPN5 W5PMR8 Q8BKX6 A0A2Y9KTD8 A0A2Y9KFV9 A0A3Q7TUJ0 A0A3Q7SEF6 J9NTC0 A0A2Y9KF75 A0A3Q7ST86 A0A2K5Q9B5 A0A2Y9HQ56 R0JWW4 F6S837 A0A3Q7MW16 A0A0B8RZ42 A0A287BD11 A0A2U3ZQP5 F1MBL6 A0A2K5EVM8 G7NPR4 A0A1U7T5V5 A0A2K5EVM1 A0A2K5EVL0 Q96Q15-2 A0A2U3WMW2 U3IWI0 H0VLT8 A0A2K5Q9U0 K9J0V7 F7F0R6 W5PMR0 A0A287B556 A0A096NRG0 A0A2J8Q5H4 A0A2K5Q9U5 A0A2R9BHL9 Q96Q15-4 G3R3L1
EC Number
2.7.11.1
Pubmed
19121390
28756777
26354079
26227816
22118469
24845553
+ More
24508170 20798317 29403074 30249741 28648823 20075255 21282665 21347285 21719571 20566863 18362917 19820115 28004739 23537049 27762356 16341006 21993624 24813606 20809919 19468303 14621295 16141072 17242355 21183079 23806337 25243066 30723633 19393038 22002653 9566925 11544179 11331269 15175154 15616553 9455477 12168954 8524286 11948212 14636577 12554878 16452507 16964243 17525332 18669648 19413330 19417104 19690332 19608861 20427287 20810650 20371770 20068231 21269460 21245168 21406692 23186163 24275569 17344846 23749191 17495919 22722832
24508170 20798317 29403074 30249741 28648823 20075255 21282665 21347285 21719571 20566863 18362917 19820115 28004739 23537049 27762356 16341006 21993624 24813606 20809919 19468303 14621295 16141072 17242355 21183079 23806337 25243066 30723633 19393038 22002653 9566925 11544179 11331269 15175154 15616553 9455477 12168954 8524286 11948212 14636577 12554878 16452507 16964243 17525332 18669648 19413330 19417104 19690332 19608861 20427287 20810650 20371770 20068231 21269460 21245168 21406692 23186163 24275569 17344846 23749191 17495919 22722832
EMBL
BABH01028615
BABH01028616
BABH01028617
NWSH01001945
PCG69606.1
PCG69605.1
+ More
KZ149897 PZC78706.1 KQ459692 KPJ20436.1 JTDY01000627 KOB76417.1 KQ459590 KPI97135.1 ODYU01004851 SOQ45151.1 RSAL01000104 RVE47354.1 AGBW02014545 OWR41458.1 KQ760382 OAD60741.1 KQ435714 KOX79166.1 KK852999 KDR12657.1 KQ434796 KZC05708.1 KQ414615 KOC68709.1 KZ288204 PBC33161.1 LBMM01000708 KMQ97679.1 KK107020 EZA62474.1 GL452737 EFN76953.1 PYGN01000433 PSN46478.1 GBYB01010851 GBYB01010852 GBYB01010853 GBYB01010854 GBYB01010856 JAG80618.1 JAG80619.1 JAG80620.1 JAG80621.1 JAG80623.1 GL434491 EFN74897.1 QOIP01000004 RLU23717.1 NNAY01000979 OXU25637.1 GL763174 EFZ20104.1 KQ978143 KYM96819.1 KQ979275 KYN22035.1 KQ982775 KYQ50767.1 KQ981522 KYN40743.1 KQ976445 KYM86128.1 ADTU01026180 ADTU01026181 GL888355 EGI62204.1 DS235012 EEB10453.1 KQ971319 KYB28836.1 LJIG01016207 KRT81293.1 GEDC01022287 JAS15011.1 GEDC01026897 JAS10401.1 GEZM01019431 JAV89636.1 APGK01019726 KB740120 ENN81315.1 KB631607 ERL84635.1 JH431789 ACPB03006723 ACPB03006724 ACPB03006725 GFTR01008998 JAW07428.1 ABLF02032469 ABLF02032472 GFXV01002049 MBW13854.1 GGMR01009303 MBY21922.1 CM004482 OCT64106.1 CM004483 OCT61503.1 AAEX03004486 AAEX03004487 KL206788 KFV85695.1 AAGW02057093 AGTP01030858 AGTP01030859 AGTP01030860 ADFV01031577 ADFV01031578 ADFV01031579 AEYP01027288 AEYP01027289 AEYP01027290 AEYP01027291 AEYP01027292 AEYP01027293 AMGL01068643 AMGL01068644 AC131788 AC132432 AK129136 AK220328 AK041264 AK047829 AK049391 AK052331 KB743005 EOB02051.1 GAMR01003557 GAMR01003556 JAB30375.1 GBZA01000616 JAG69152.1 AEMK02000017 DQIR01031496 DQIR01166796 DQIR01199951 HCZ86971.1 CM001272 EHH31463.1 AF186377 AB061371 AY014957 AF395444 AC092287 AB007881 U32581 ADON01065093 AAKN02007594 AAKN02007595 GABZ01003339 JAA50186.1 AHZZ02018445 AHZZ02018446 NBAG03000076 PNI91496.1 CABD030099509 CABD030099510 CABD030099511 CABD030099512 CABD030099513 CABD030099514 CABD030099515 CABD030099516 CABD030099517 CABD030099518
KZ149897 PZC78706.1 KQ459692 KPJ20436.1 JTDY01000627 KOB76417.1 KQ459590 KPI97135.1 ODYU01004851 SOQ45151.1 RSAL01000104 RVE47354.1 AGBW02014545 OWR41458.1 KQ760382 OAD60741.1 KQ435714 KOX79166.1 KK852999 KDR12657.1 KQ434796 KZC05708.1 KQ414615 KOC68709.1 KZ288204 PBC33161.1 LBMM01000708 KMQ97679.1 KK107020 EZA62474.1 GL452737 EFN76953.1 PYGN01000433 PSN46478.1 GBYB01010851 GBYB01010852 GBYB01010853 GBYB01010854 GBYB01010856 JAG80618.1 JAG80619.1 JAG80620.1 JAG80621.1 JAG80623.1 GL434491 EFN74897.1 QOIP01000004 RLU23717.1 NNAY01000979 OXU25637.1 GL763174 EFZ20104.1 KQ978143 KYM96819.1 KQ979275 KYN22035.1 KQ982775 KYQ50767.1 KQ981522 KYN40743.1 KQ976445 KYM86128.1 ADTU01026180 ADTU01026181 GL888355 EGI62204.1 DS235012 EEB10453.1 KQ971319 KYB28836.1 LJIG01016207 KRT81293.1 GEDC01022287 JAS15011.1 GEDC01026897 JAS10401.1 GEZM01019431 JAV89636.1 APGK01019726 KB740120 ENN81315.1 KB631607 ERL84635.1 JH431789 ACPB03006723 ACPB03006724 ACPB03006725 GFTR01008998 JAW07428.1 ABLF02032469 ABLF02032472 GFXV01002049 MBW13854.1 GGMR01009303 MBY21922.1 CM004482 OCT64106.1 CM004483 OCT61503.1 AAEX03004486 AAEX03004487 KL206788 KFV85695.1 AAGW02057093 AGTP01030858 AGTP01030859 AGTP01030860 ADFV01031577 ADFV01031578 ADFV01031579 AEYP01027288 AEYP01027289 AEYP01027290 AEYP01027291 AEYP01027292 AEYP01027293 AMGL01068643 AMGL01068644 AC131788 AC132432 AK129136 AK220328 AK041264 AK047829 AK049391 AK052331 KB743005 EOB02051.1 GAMR01003557 GAMR01003556 JAB30375.1 GBZA01000616 JAG69152.1 AEMK02000017 DQIR01031496 DQIR01166796 DQIR01199951 HCZ86971.1 CM001272 EHH31463.1 AF186377 AB061371 AY014957 AF395444 AC092287 AB007881 U32581 ADON01065093 AAKN02007594 AAKN02007595 GABZ01003339 JAA50186.1 AHZZ02018445 AHZZ02018446 NBAG03000076 PNI91496.1 CABD030099509 CABD030099510 CABD030099511 CABD030099512 CABD030099513 CABD030099514 CABD030099515 CABD030099516 CABD030099517 CABD030099518
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000053268
UP000283053
+ More
UP000007151 UP000053105 UP000027135 UP000076502 UP000053825 UP000005203 UP000242457 UP000036403 UP000053097 UP000008237 UP000245037 UP000000311 UP000279307 UP000215335 UP000002358 UP000078542 UP000078492 UP000075809 UP000078541 UP000078540 UP000005205 UP000007755 UP000009046 UP000007266 UP000019118 UP000030742 UP000015103 UP000007819 UP000186698 UP000002254 UP000053584 UP000007646 UP000286642 UP000001811 UP000005215 UP000001073 UP000261680 UP000291021 UP000000715 UP000002356 UP000000589 UP000248482 UP000286640 UP000233040 UP000248481 UP000008225 UP000286641 UP000008227 UP000245340 UP000009136 UP000233020 UP000189704 UP000005640 UP000016666 UP000005447 UP000002280 UP000028761 UP000240080 UP000001519
UP000007151 UP000053105 UP000027135 UP000076502 UP000053825 UP000005203 UP000242457 UP000036403 UP000053097 UP000008237 UP000245037 UP000000311 UP000279307 UP000215335 UP000002358 UP000078542 UP000078492 UP000075809 UP000078541 UP000078540 UP000005205 UP000007755 UP000009046 UP000007266 UP000019118 UP000030742 UP000015103 UP000007819 UP000186698 UP000002254 UP000053584 UP000007646 UP000286642 UP000001811 UP000005215 UP000001073 UP000261680 UP000291021 UP000000715 UP000002356 UP000000589 UP000248482 UP000286640 UP000233040 UP000248481 UP000008225 UP000286641 UP000008227 UP000245340 UP000009136 UP000233020 UP000189704 UP000005640 UP000016666 UP000005447 UP000002280 UP000028761 UP000240080 UP000001519
Interpro
Gene 3D
ProteinModelPortal
H9JR19
A0A2A4JCX2
A0A2A4JC06
A0A2W1C160
A0A0N1PK13
A0A0L7LLT8
+ More
A0A194Q159 A0A2H1VWC4 A0A3S2NCA3 A0A212EJ17 A0A310SKL6 A0A0M9A9D8 A0A067R3Y6 A0A154P395 A0A0L7RD16 A0A087ZS52 A0A2A3EQ41 A0A0J7L5B8 A0A026X4W6 E2C545 A0A2P8YQE7 A0A0C9RCL1 E1ZVC1 A0A3L8DTG4 A0A232F5L9 K7J769 E9IH54 A0A195C7G7 A0A151JAE5 A0A151WSJ2 A0A195FK18 A0A151I5P1 A0A158NTX1 F4WU96 E0VAP7 A0A139WLQ5 A0A0T6B2K1 A0A1B6CNV0 A0A1B6CAC4 A0A1Y1N066 N6UHU4 U4U3T0 T1J1T4 T1I2T7 A0A224XCD0 J9KB53 A0A2H8TIB2 A0A2S2NXY1 A0A1L8EXP0 A0A1L8EQ71 F1PBU5 A0A093I1J3 G3TL75 A0A3Q7Y545 A0A3Q7XMV0 G1TC15 I3MMW0 A0A287CYX9 G1R0C7 A0A384CK10 A0A3Q7XEQ5 M3YPN5 W5PMR8 Q8BKX6 A0A2Y9KTD8 A0A2Y9KFV9 A0A3Q7TUJ0 A0A3Q7SEF6 J9NTC0 A0A2Y9KF75 A0A3Q7ST86 A0A2K5Q9B5 A0A2Y9HQ56 R0JWW4 F6S837 A0A3Q7MW16 A0A0B8RZ42 A0A287BD11 A0A2U3ZQP5 F1MBL6 A0A2K5EVM8 G7NPR4 A0A1U7T5V5 A0A2K5EVM1 A0A2K5EVL0 Q96Q15-2 A0A2U3WMW2 U3IWI0 H0VLT8 A0A2K5Q9U0 K9J0V7 F7F0R6 W5PMR0 A0A287B556 A0A096NRG0 A0A2J8Q5H4 A0A2K5Q9U5 A0A2R9BHL9 Q96Q15-4 G3R3L1
A0A194Q159 A0A2H1VWC4 A0A3S2NCA3 A0A212EJ17 A0A310SKL6 A0A0M9A9D8 A0A067R3Y6 A0A154P395 A0A0L7RD16 A0A087ZS52 A0A2A3EQ41 A0A0J7L5B8 A0A026X4W6 E2C545 A0A2P8YQE7 A0A0C9RCL1 E1ZVC1 A0A3L8DTG4 A0A232F5L9 K7J769 E9IH54 A0A195C7G7 A0A151JAE5 A0A151WSJ2 A0A195FK18 A0A151I5P1 A0A158NTX1 F4WU96 E0VAP7 A0A139WLQ5 A0A0T6B2K1 A0A1B6CNV0 A0A1B6CAC4 A0A1Y1N066 N6UHU4 U4U3T0 T1J1T4 T1I2T7 A0A224XCD0 J9KB53 A0A2H8TIB2 A0A2S2NXY1 A0A1L8EXP0 A0A1L8EQ71 F1PBU5 A0A093I1J3 G3TL75 A0A3Q7Y545 A0A3Q7XMV0 G1TC15 I3MMW0 A0A287CYX9 G1R0C7 A0A384CK10 A0A3Q7XEQ5 M3YPN5 W5PMR8 Q8BKX6 A0A2Y9KTD8 A0A2Y9KFV9 A0A3Q7TUJ0 A0A3Q7SEF6 J9NTC0 A0A2Y9KF75 A0A3Q7ST86 A0A2K5Q9B5 A0A2Y9HQ56 R0JWW4 F6S837 A0A3Q7MW16 A0A0B8RZ42 A0A287BD11 A0A2U3ZQP5 F1MBL6 A0A2K5EVM8 G7NPR4 A0A1U7T5V5 A0A2K5EVM1 A0A2K5EVL0 Q96Q15-2 A0A2U3WMW2 U3IWI0 H0VLT8 A0A2K5Q9U0 K9J0V7 F7F0R6 W5PMR0 A0A287B556 A0A096NRG0 A0A2J8Q5H4 A0A2K5Q9U5 A0A2R9BHL9 Q96Q15-4 G3R3L1
Ontologies
GO
GO:0016301
GO:0005524
GO:0000184
GO:0004674
GO:0005634
GO:0046854
GO:0005737
GO:0042162
GO:0018105
GO:2001020
GO:0046777
GO:0005654
GO:0032204
GO:0016021
GO:0006281
GO:0046872
GO:0005829
GO:0004672
GO:0003723
GO:0006406
GO:0008270
GO:0003824
GO:0009982
GO:0006744
GO:0016627
GO:0030145
GO:0015923
GO:0006260
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
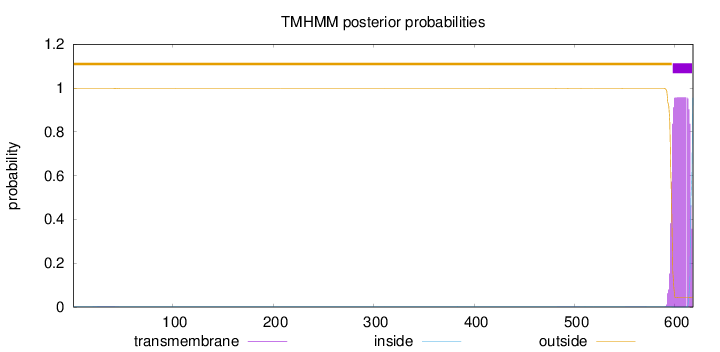
Length:
618
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.01563
Exp number, first 60 AAs:
0.03413
Total prob of N-in:
0.00315
outside
1 - 597
TMhelix
598 - 617
inside
618 - 618
Population Genetic Test Statistics
Pi
244.206059
Theta
175.126662
Tajima's D
1.044368
CLR
0.083448
CSRT
0.67591620418979
Interpretation
Uncertain
