Gene
KWMTBOMO06353 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011984
Annotation
Alanine_aminotransferase_1_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.481
Sequence
CDS
ATGAAGTTAGAAAAGTGTTTAACTGAGGAGAACATTAATCAGAATTTACTGAATATCCAGTATGCGGTCCGTGGACCTATCTTGCAGAGGGCTTTGCAGATCGAAAGAGAGTTAGTCAAGGGGGTTCCGAAGCGCTTCGAGCGTGTGGTGAGAGCTAACTGCGGCGACTGTCACGCTCTCGGACAGAAACCCATCACATTCATAAGACAGGTATTAGCCCTAGTGTCATTCCCGGAACTGATCTCGTCTCGTCTTGATATCCCTCAAGACGTGAAGGACAGAGCCAAAGAGCTACTCGACGACTGCACGAAAGGTTCAGTGGGATCGTATTCTCCATCCAACGGTCTCAACTTGGTGCGATCTCGTGTCGCACAGTACATCACGAACAGAGACGGTGTGCCTTCCAACCCTGACGATATTTACCTGGGATCTGGCGCCTCTGATGTTATTAAATCAGTCCTAACTCTGTTTGTTGAGGATGTGGGAGGAAAGCCCCCCGCTGTGATGGTGCCGATCCCTCAGTACCCATTGTTTTCGGGCACTCTCTCAGAGCTAGGTGTTCGACAGGTCGACTATTACCTTGACGAGGAACACGACTGGGCTCTTCAGATCTCAGAGCTAGAAAGGAGCTGGCGAGAGGCCAGTATTGAGAGTAATGTGCGGGCTCTTGTTGTGATCAACCCTGGAAATCCTACAGGACAGGTGTTAACCCGCGATAATATTGAACAAATCATCAAATTTGCTTACGAGCGTAACTTGTTCATTCTGGCTGACGAAGTATACCAGGAGAACATCGTGAGCAAGCCGTTTCATTCCTTTAAAAAGGTAATGTTCGAAATGGGTGCACCATACTCCCGCATGGAATTGGCTAGTTTCTTGACGGCCTCAAAAGGCTGGGCAGCTGAGTGTGGCTTGAGGTCGGGATACGTGGAGCTACTGCATCTCCAACCATCAGTGCAGAAAACGTTCAATACAATGAGGTCTGTCATGCAGTGCCCCTCGGTTTTGGGACAGTGCATTTTGGATTGCGTGATGAAGCCTCCGTCTCCCGGAAAACCCTCACACAATCAGTTCATCAATGAAATTAGAGACATTCATCAAGTATTAAAGGAACGCACTGCAACCGCTTACAAAACCTTCAACTCAATACCAGGATATTTCTGCAATCCAATCGACGGTTCGATGTTCGCATATCCTCGTATAGAAATTCCAGTAGAAGCCCAATTAGAAGCAAAAAAACTGAACATGAGTCCTGATGAGTTCTACTGCCTTAGGCTGTTAGAGGAAACTGGAGTGTGTGTGATCCCGGGTACGGGATTCGGGCAGCTTCCAGGATCATTCCATTTCAGAACTACGATACTCCACCCCAAGGACGAATTCCAGTACATGATGGACTCTATAAGACGTTTCCACTTGAACTTCATGCAGTTATATCCTTAG
Protein
MKLEKCLTEENINQNLLNIQYAVRGPILQRALQIERELVKGVPKRFERVVRANCGDCHALGQKPITFIRQVLALVSFPELISSRLDIPQDVKDRAKELLDDCTKGSVGSYSPSNGLNLVRSRVAQYITNRDGVPSNPDDIYLGSGASDVIKSVLTLFVEDVGGKPPAVMVPIPQYPLFSGTLSELGVRQVDYYLDEEHDWALQISELERSWREASIESNVRALVVINPGNPTGQVLTRDNIEQIIKFAYERNLFILADEVYQENIVSKPFHSFKKVMFEMGAPYSRMELASFLTASKGWAAECGLRSGYVELLHLQPSVQKTFNTMRSVMQCPSVLGQCILDCVMKPPSPGKPSHNQFINEIRDIHQVLKERTATAYKTFNSIPGYFCNPIDGSMFAYPRIEIPVEAQLEAKKLNMSPDEFYCLRLLEETGVCVIPGTGFGQLPGSFHFRTTILHPKDEFQYMMDSIRRFHLNFMQLYP
Summary
Uniprot
H9JR24
A0A2H1V0Y9
A0A2A4JWS8
A0A2W1BUG6
A0A194PUX5
A0A0N1IH94
+ More
A0A1D2NAV9 A0A0G2SJ75 J9JZJ2 A0A1B6CHN5 A0A2L2XWP5 A0A2A3E2I9 V9IKQ3 A0A2L2XYY3 A0A2L2XVU2 A0A154PAC0 A0A2P6L828 A0A069DUE3 A0A195CWX5 A0A087ZNZ2 A0A310SBG8 A0A0L7RAR0 V5HPG4 A0A1B6ENS5 A0A2J7R9U4 A0A023F5A9 E9J4Y5 A0A1S6KZB9 A0A1E1X8F5 A0A151X0M4 A0A1W4XJ77 A0A1I9WLI3 K7IY19 A0A023FJZ7 A0A2R5LAM2 A0A023GFF6 X5DCE6 A0A0K0K0N7 A0A067QSN5 A0A026WG43 E0VVX2 L7LZX8 A0A131XH19 A0A182GT39 A0A131Z6Z0 A0A224YMM7 A0A1Z5KVG2 A0A0L0BRJ7 A0A1L8E9U9 A0A1L8EA18 A0A2Z5RE46 A0A1L8EAA0 N6TE08 A0A1L8EAN2 A0A0A9WUZ7 E1ZV08 A0A158RBL9 E2B482 A0A2A4JWU3 A0A0K8TPJ7 A0A0P5GHV1 A0A151JVJ7 A0A158NM17 A0A1I8P9H3 A0A0P4VK84 T1HPT2 Q16UK8 A0A0N4TWH6 A0A1S4FNL0 A0A194PVV1 A0A0P5T0B7 A0A336KUI2 A0A0P5SSK9 A0A0P4ZHR5 A0A0P5K8W3 A0A0P5KUP1 A0A2W1C0T3 R4WQ57 A0A1J1HTD5 A8WW42 A0A0P5UE15 A0A0P5SJD0 A0A0N8CEP4 A0A0P5JCG1 A0A0P5ECD3 A0A0P5JLY7 U5ER68 A0A2K6VPU4 A0A0P6I2A2 A0A0N8A3R4 K1QAF3 E9HAN7 A0A1L8E2L8 A0A0N1PJH1 A0A1L8E2C5 B5DLB2 A0A0K8WBV8 A0A3B4DT52 F7AQB9 A0A2G5VEB3
A0A1D2NAV9 A0A0G2SJ75 J9JZJ2 A0A1B6CHN5 A0A2L2XWP5 A0A2A3E2I9 V9IKQ3 A0A2L2XYY3 A0A2L2XVU2 A0A154PAC0 A0A2P6L828 A0A069DUE3 A0A195CWX5 A0A087ZNZ2 A0A310SBG8 A0A0L7RAR0 V5HPG4 A0A1B6ENS5 A0A2J7R9U4 A0A023F5A9 E9J4Y5 A0A1S6KZB9 A0A1E1X8F5 A0A151X0M4 A0A1W4XJ77 A0A1I9WLI3 K7IY19 A0A023FJZ7 A0A2R5LAM2 A0A023GFF6 X5DCE6 A0A0K0K0N7 A0A067QSN5 A0A026WG43 E0VVX2 L7LZX8 A0A131XH19 A0A182GT39 A0A131Z6Z0 A0A224YMM7 A0A1Z5KVG2 A0A0L0BRJ7 A0A1L8E9U9 A0A1L8EA18 A0A2Z5RE46 A0A1L8EAA0 N6TE08 A0A1L8EAN2 A0A0A9WUZ7 E1ZV08 A0A158RBL9 E2B482 A0A2A4JWU3 A0A0K8TPJ7 A0A0P5GHV1 A0A151JVJ7 A0A158NM17 A0A1I8P9H3 A0A0P4VK84 T1HPT2 Q16UK8 A0A0N4TWH6 A0A1S4FNL0 A0A194PVV1 A0A0P5T0B7 A0A336KUI2 A0A0P5SSK9 A0A0P4ZHR5 A0A0P5K8W3 A0A0P5KUP1 A0A2W1C0T3 R4WQ57 A0A1J1HTD5 A8WW42 A0A0P5UE15 A0A0P5SJD0 A0A0N8CEP4 A0A0P5JCG1 A0A0P5ECD3 A0A0P5JLY7 U5ER68 A0A2K6VPU4 A0A0P6I2A2 A0A0N8A3R4 K1QAF3 E9HAN7 A0A1L8E2L8 A0A0N1PJH1 A0A1L8E2C5 B5DLB2 A0A0K8WBV8 A0A3B4DT52 F7AQB9 A0A2G5VEB3
Pubmed
19121390
28756777
26354079
27289101
26561354
26334808
+ More
25765539 25474469 21282665 24330026 28503490 27538518 20075255 17885136 24845553 24508170 30249741 20566863 25576852 28049606 26483478 26830274 28797301 28528879 26108605 26760975 23537049 25401762 26823975 20798317 26369729 21347285 27129103 17510324 23691247 14624247 22992520 21292972 15632085 17994087 23185243 17495919
25765539 25474469 21282665 24330026 28503490 27538518 20075255 17885136 24845553 24508170 30249741 20566863 25576852 28049606 26483478 26830274 28797301 28528879 26108605 26760975 23537049 25401762 26823975 20798317 26369729 21347285 27129103 17510324 23691247 14624247 22992520 21292972 15632085 17994087 23185243 17495919
EMBL
BABH01028651
ODYU01000172
SOQ34507.1
NWSH01000529
PCG75860.1
KZ149897
+ More
PZC78722.1 KQ459590 KPI97122.1 KQ459692 KPJ20450.1 LJIJ01000113 ODN02389.1 KJ802126 AJP75146.1 ABLF02024379 GEDC01024415 JAS12883.1 IAAA01005953 LAA00217.1 KZ288443 PBC25694.1 JR049220 AEY60899.1 IAAA01005954 LAA00220.1 IAAA01005955 LAA00223.1 KQ434857 KZC08802.1 MWRG01001106 PRD34734.1 GBGD01001314 JAC87575.1 KQ977171 KYN05173.1 KQ771982 OAD52523.1 KQ414618 KOC67910.1 GANP01008735 JAB75733.1 GECZ01030202 GECZ01029764 GECZ01016955 JAS39567.1 JAS40005.1 JAS52814.1 NEVH01006580 PNF37605.1 GBBI01002423 JAC16289.1 GL768121 EFZ12113.1 KC223169 AQT27072.1 GFAC01003646 JAT95542.1 KQ982610 KYQ53914.1 KU932367 APA34003.1 GBBK01002316 JAC22166.1 GGLE01002427 MBY06553.1 GBBM01003720 JAC31698.1 KJ000066 AHW58022.1 KK852999 KDR12685.1 KK107238 QOIP01000007 EZA54611.1 RLU20662.1 DS235815 EEB17528.1 GACK01007887 JAA57147.1 GEFH01003615 JAP64966.1 JXUM01086158 JXUM01086159 JXUM01086160 JXUM01086161 GEDV01001809 JAP86748.1 GFPF01005853 MAA16999.1 GFJQ02007891 JAV99078.1 JRES01001467 KNC22662.1 GFDG01003288 JAV15511.1 GFDG01003340 JAV15459.1 FX983741 BAX07240.1 GFDG01003132 JAV15667.1 APGK01033055 KB740860 KB632287 ENN78579.1 ERL91463.1 GFDG01003133 JAV15666.1 GBHO01033266 GBRD01003195 GDHC01004408 JAG10338.1 JAG62626.1 JAQ14221.1 GL434335 EFN75006.1 UYYF01004314 VDN02184.1 GL445515 EFN89499.1 PCG75862.1 GDAI01001530 JAI16073.1 GDIQ01240448 JAK11277.1 KQ981688 KYN37925.1 ADTU01020144 GDKW01001133 JAI55462.1 ACPB03001903 CH477620 EAT38206.1 UZAD01013366 VDN94402.1 KPI97123.1 GDIP01213840 GDIP01212940 GDIP01212939 GDIP01148492 GDIQ01243195 GDIQ01241842 GDIQ01224508 GDIP01142444 GDIP01142390 GDIP01135292 GDIP01135291 GDIP01134052 JAK08530.1 JAL68423.1 UFQS01000956 UFQT01000956 SSX07936.1 SSX28170.1 GDIP01139094 JAL64620.1 GDIP01213839 GDIP01212938 GDIP01153627 GDIP01152835 GDIP01151035 GDIQ01241843 GDIP01142443 GDIP01140034 GDIP01135294 GDIP01134053 GDIP01134050 JAJ09563.1 JAK09882.1 GDIQ01188810 JAK62915.1 GDIQ01204752 JAK46973.1 PZC78720.1 AK417826 BAN21041.1 CVRI01000020 CRK91319.1 HE600906 CAP24851.1 GDIP01212123 GDIP01131547 JAL72167.1 GDIP01222728 GDIP01213838 GDIP01212937 GDIP01151037 GDIP01148494 GDIP01148491 GDIQ01244394 GDIQ01243196 GDIQ01240449 GDIQ01225401 GDIQ01190412 GDIP01142442 GDIP01140033 GDIP01139093 GDIP01139092 GDIP01135293 GDIQ01037497 JAK07331.1 JAL64622.1 GDIP01139095 GDIP01139090 JAL64619.1 GDIQ01207604 JAK44121.1 GDIP01222729 GDIP01163691 GDIP01151097 GDIP01151036 GDIP01148493 GDIQ01244393 GDIQ01243194 GDIQ01240391 GDIQ01187358 GDIP01140036 GDIP01140035 GDIP01140032 GDIP01139091 GDIP01134051 GDIP01131548 JAJ74909.1 JAK07332.1 GDIQ01203005 JAK48720.1 GANO01002941 JAB56930.1 CMVM020000044 GDIQ01011648 JAN83089.1 GDIP01185426 GDIP01185425 JAJ37977.1 JH816600 EKC33627.1 GL732612 EFX71233.1 GFDF01001215 JAV12869.1 KPJ20449.1 GFDF01001216 JAV12868.1 CH379063 EDY72311.2 KRT06146.1 GDHF01022472 GDHF01003780 JAI29842.1 JAI48534.1 PDUG01000001 PIC50093.1
PZC78722.1 KQ459590 KPI97122.1 KQ459692 KPJ20450.1 LJIJ01000113 ODN02389.1 KJ802126 AJP75146.1 ABLF02024379 GEDC01024415 JAS12883.1 IAAA01005953 LAA00217.1 KZ288443 PBC25694.1 JR049220 AEY60899.1 IAAA01005954 LAA00220.1 IAAA01005955 LAA00223.1 KQ434857 KZC08802.1 MWRG01001106 PRD34734.1 GBGD01001314 JAC87575.1 KQ977171 KYN05173.1 KQ771982 OAD52523.1 KQ414618 KOC67910.1 GANP01008735 JAB75733.1 GECZ01030202 GECZ01029764 GECZ01016955 JAS39567.1 JAS40005.1 JAS52814.1 NEVH01006580 PNF37605.1 GBBI01002423 JAC16289.1 GL768121 EFZ12113.1 KC223169 AQT27072.1 GFAC01003646 JAT95542.1 KQ982610 KYQ53914.1 KU932367 APA34003.1 GBBK01002316 JAC22166.1 GGLE01002427 MBY06553.1 GBBM01003720 JAC31698.1 KJ000066 AHW58022.1 KK852999 KDR12685.1 KK107238 QOIP01000007 EZA54611.1 RLU20662.1 DS235815 EEB17528.1 GACK01007887 JAA57147.1 GEFH01003615 JAP64966.1 JXUM01086158 JXUM01086159 JXUM01086160 JXUM01086161 GEDV01001809 JAP86748.1 GFPF01005853 MAA16999.1 GFJQ02007891 JAV99078.1 JRES01001467 KNC22662.1 GFDG01003288 JAV15511.1 GFDG01003340 JAV15459.1 FX983741 BAX07240.1 GFDG01003132 JAV15667.1 APGK01033055 KB740860 KB632287 ENN78579.1 ERL91463.1 GFDG01003133 JAV15666.1 GBHO01033266 GBRD01003195 GDHC01004408 JAG10338.1 JAG62626.1 JAQ14221.1 GL434335 EFN75006.1 UYYF01004314 VDN02184.1 GL445515 EFN89499.1 PCG75862.1 GDAI01001530 JAI16073.1 GDIQ01240448 JAK11277.1 KQ981688 KYN37925.1 ADTU01020144 GDKW01001133 JAI55462.1 ACPB03001903 CH477620 EAT38206.1 UZAD01013366 VDN94402.1 KPI97123.1 GDIP01213840 GDIP01212940 GDIP01212939 GDIP01148492 GDIQ01243195 GDIQ01241842 GDIQ01224508 GDIP01142444 GDIP01142390 GDIP01135292 GDIP01135291 GDIP01134052 JAK08530.1 JAL68423.1 UFQS01000956 UFQT01000956 SSX07936.1 SSX28170.1 GDIP01139094 JAL64620.1 GDIP01213839 GDIP01212938 GDIP01153627 GDIP01152835 GDIP01151035 GDIQ01241843 GDIP01142443 GDIP01140034 GDIP01135294 GDIP01134053 GDIP01134050 JAJ09563.1 JAK09882.1 GDIQ01188810 JAK62915.1 GDIQ01204752 JAK46973.1 PZC78720.1 AK417826 BAN21041.1 CVRI01000020 CRK91319.1 HE600906 CAP24851.1 GDIP01212123 GDIP01131547 JAL72167.1 GDIP01222728 GDIP01213838 GDIP01212937 GDIP01151037 GDIP01148494 GDIP01148491 GDIQ01244394 GDIQ01243196 GDIQ01240449 GDIQ01225401 GDIQ01190412 GDIP01142442 GDIP01140033 GDIP01139093 GDIP01139092 GDIP01135293 GDIQ01037497 JAK07331.1 JAL64622.1 GDIP01139095 GDIP01139090 JAL64619.1 GDIQ01207604 JAK44121.1 GDIP01222729 GDIP01163691 GDIP01151097 GDIP01151036 GDIP01148493 GDIQ01244393 GDIQ01243194 GDIQ01240391 GDIQ01187358 GDIP01140036 GDIP01140035 GDIP01140032 GDIP01139091 GDIP01134051 GDIP01131548 JAJ74909.1 JAK07332.1 GDIQ01203005 JAK48720.1 GANO01002941 JAB56930.1 CMVM020000044 GDIQ01011648 JAN83089.1 GDIP01185426 GDIP01185425 JAJ37977.1 JH816600 EKC33627.1 GL732612 EFX71233.1 GFDF01001215 JAV12869.1 KPJ20449.1 GFDF01001216 JAV12868.1 CH379063 EDY72311.2 KRT06146.1 GDHF01022472 GDHF01003780 JAI29842.1 JAI48534.1 PDUG01000001 PIC50093.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000094527
UP000007819
+ More
UP000242457 UP000076502 UP000078542 UP000005203 UP000053825 UP000235965 UP000075809 UP000192223 UP000002358 UP000006672 UP000027135 UP000053097 UP000279307 UP000009046 UP000069940 UP000037069 UP000019118 UP000030742 UP000000311 UP000046394 UP000276776 UP000008237 UP000078541 UP000005205 UP000095300 UP000015103 UP000008820 UP000038020 UP000278627 UP000183832 UP000008549 UP000024404 UP000005408 UP000000305 UP000001819 UP000261440 UP000002280 UP000230233
UP000242457 UP000076502 UP000078542 UP000005203 UP000053825 UP000235965 UP000075809 UP000192223 UP000002358 UP000006672 UP000027135 UP000053097 UP000279307 UP000009046 UP000069940 UP000037069 UP000019118 UP000030742 UP000000311 UP000046394 UP000276776 UP000008237 UP000078541 UP000005205 UP000095300 UP000015103 UP000008820 UP000038020 UP000278627 UP000183832 UP000008549 UP000024404 UP000005408 UP000000305 UP000001819 UP000261440 UP000002280 UP000230233
Pfam
PF00155 Aminotran_1_2
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
H9JR24
A0A2H1V0Y9
A0A2A4JWS8
A0A2W1BUG6
A0A194PUX5
A0A0N1IH94
+ More
A0A1D2NAV9 A0A0G2SJ75 J9JZJ2 A0A1B6CHN5 A0A2L2XWP5 A0A2A3E2I9 V9IKQ3 A0A2L2XYY3 A0A2L2XVU2 A0A154PAC0 A0A2P6L828 A0A069DUE3 A0A195CWX5 A0A087ZNZ2 A0A310SBG8 A0A0L7RAR0 V5HPG4 A0A1B6ENS5 A0A2J7R9U4 A0A023F5A9 E9J4Y5 A0A1S6KZB9 A0A1E1X8F5 A0A151X0M4 A0A1W4XJ77 A0A1I9WLI3 K7IY19 A0A023FJZ7 A0A2R5LAM2 A0A023GFF6 X5DCE6 A0A0K0K0N7 A0A067QSN5 A0A026WG43 E0VVX2 L7LZX8 A0A131XH19 A0A182GT39 A0A131Z6Z0 A0A224YMM7 A0A1Z5KVG2 A0A0L0BRJ7 A0A1L8E9U9 A0A1L8EA18 A0A2Z5RE46 A0A1L8EAA0 N6TE08 A0A1L8EAN2 A0A0A9WUZ7 E1ZV08 A0A158RBL9 E2B482 A0A2A4JWU3 A0A0K8TPJ7 A0A0P5GHV1 A0A151JVJ7 A0A158NM17 A0A1I8P9H3 A0A0P4VK84 T1HPT2 Q16UK8 A0A0N4TWH6 A0A1S4FNL0 A0A194PVV1 A0A0P5T0B7 A0A336KUI2 A0A0P5SSK9 A0A0P4ZHR5 A0A0P5K8W3 A0A0P5KUP1 A0A2W1C0T3 R4WQ57 A0A1J1HTD5 A8WW42 A0A0P5UE15 A0A0P5SJD0 A0A0N8CEP4 A0A0P5JCG1 A0A0P5ECD3 A0A0P5JLY7 U5ER68 A0A2K6VPU4 A0A0P6I2A2 A0A0N8A3R4 K1QAF3 E9HAN7 A0A1L8E2L8 A0A0N1PJH1 A0A1L8E2C5 B5DLB2 A0A0K8WBV8 A0A3B4DT52 F7AQB9 A0A2G5VEB3
A0A1D2NAV9 A0A0G2SJ75 J9JZJ2 A0A1B6CHN5 A0A2L2XWP5 A0A2A3E2I9 V9IKQ3 A0A2L2XYY3 A0A2L2XVU2 A0A154PAC0 A0A2P6L828 A0A069DUE3 A0A195CWX5 A0A087ZNZ2 A0A310SBG8 A0A0L7RAR0 V5HPG4 A0A1B6ENS5 A0A2J7R9U4 A0A023F5A9 E9J4Y5 A0A1S6KZB9 A0A1E1X8F5 A0A151X0M4 A0A1W4XJ77 A0A1I9WLI3 K7IY19 A0A023FJZ7 A0A2R5LAM2 A0A023GFF6 X5DCE6 A0A0K0K0N7 A0A067QSN5 A0A026WG43 E0VVX2 L7LZX8 A0A131XH19 A0A182GT39 A0A131Z6Z0 A0A224YMM7 A0A1Z5KVG2 A0A0L0BRJ7 A0A1L8E9U9 A0A1L8EA18 A0A2Z5RE46 A0A1L8EAA0 N6TE08 A0A1L8EAN2 A0A0A9WUZ7 E1ZV08 A0A158RBL9 E2B482 A0A2A4JWU3 A0A0K8TPJ7 A0A0P5GHV1 A0A151JVJ7 A0A158NM17 A0A1I8P9H3 A0A0P4VK84 T1HPT2 Q16UK8 A0A0N4TWH6 A0A1S4FNL0 A0A194PVV1 A0A0P5T0B7 A0A336KUI2 A0A0P5SSK9 A0A0P4ZHR5 A0A0P5K8W3 A0A0P5KUP1 A0A2W1C0T3 R4WQ57 A0A1J1HTD5 A8WW42 A0A0P5UE15 A0A0P5SJD0 A0A0N8CEP4 A0A0P5JCG1 A0A0P5ECD3 A0A0P5JLY7 U5ER68 A0A2K6VPU4 A0A0P6I2A2 A0A0N8A3R4 K1QAF3 E9HAN7 A0A1L8E2L8 A0A0N1PJH1 A0A1L8E2C5 B5DLB2 A0A0K8WBV8 A0A3B4DT52 F7AQB9 A0A2G5VEB3
PDB
3IHJ
E-value=8.68627e-136,
Score=1240
Ontologies
PATHWAY
00220
Arginine biosynthesis - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
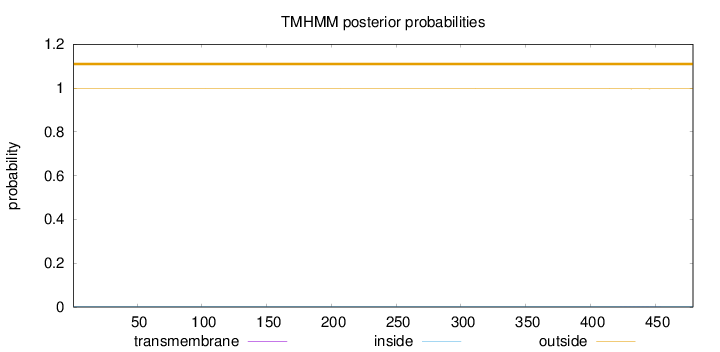
Topology
Length:
479
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01204
Exp number, first 60 AAs:
0.00043
Total prob of N-in:
0.00347
outside
1 - 479
Population Genetic Test Statistics
Pi
256.600383
Theta
168.253361
Tajima's D
1.208453
CLR
0.495748
CSRT
0.713964301784911
Interpretation
Uncertain
