Gene
KWMTBOMO06349
Pre Gene Modal
BGIBMGA011910
Annotation
PREDICTED:_Kv_channel-interacting_protein_4-like_[Amyelois_transitella]
Full name
Kv channel-interacting protein 4
Alternative Name
A-type potassium channel modulatory protein 4
Calsenilin-like protein
Potassium channel-interacting protein 4
Calsenilin-like protein
Potassium channel-interacting protein 4
Location in the cell
Cytoplasmic Reliability : 1.64
Sequence
CDS
ATGTACCGCGGGTTCAAACAGGAGTGTCCGTCGGGGGCAGTGGACGAGGAAGCCTTCAAGAACATATTCTGTCAGTTCTTCCCGCTCGGCGACGCCACGCAGTACGCGCACTACGTCTTCAACACGATCAAGCACAAGCAGAGCGGGAGGGTCAACTTCGAGGAGTTCCTGGGCATCCTGTCCCGAGTGGCACGCGGCTCCGTGCAAGAGAAGCTGTCCTGGGTGTTCGCGCTCTACGACGTCGACGGGGACGGCCGCATCTCCCGCGCCGAGATGCTGTCCGTGGTGCAAGCTATATACGAGCTGCTGGGACGAGCCGTGGCCCCCCCGGCGCACCCCACTGCCGCCAAGGACCACGTCGACAGAATCTTCCATTTGATCGACACGAACGCAGACGGCGTAGTGACTCCGGACGAGCTGGCGCGCTGGTGCAGCCGGGACCCCGCGCTGCTGCGGTCCCTGGATACCCTGGACACCGTGTTTTGTTGTGAAGGTGAAGCGATACCTTCCAGTTCAAAGGAAGCCGACGTGCCCGGCGCGACGCCCGCCAGAGTCACCGCCGTCATAACAGAAGAAGCATCTCTGAAGAAAGTAAAACGTCGGAACCGACGCTCTAGCACGCGCCGGTGA
Protein
MYRGFKQECPSGAVDEEAFKNIFCQFFPLGDATQYAHYVFNTIKHKQSGRVNFEEFLGILSRVARGSVQEKLSWVFALYDVDGDGRISRAEMLSVVQAIYELLGRAVAPPAHPTAAKDHVDRIFHLIDTNADGVVTPDELARWCSRDPALLRSLDTLDTVFCCEGEAIPSSSKEADVPGATPARVTAVITEEASLKKVKRRNRRSSTRR
Summary
Description
Regulatory subunit of Kv4/D (Shal)-type voltage-gated rapidly inactivating A-type potassium channels, such as KCND2/Kv4.2 and KCND3/Kv4.3 (PubMed:19109250). Modulates channel expression at the cell membrane, gating characteristics, inactivation kinetics and rate of recovery from inactivation in a calcium-dependent and isoform-specific manner.
Regulatory subunit of Kv4/D (Shal)-type voltage-gated rapidly inactivating A-type potassium channels. Modulates KCND2 channel density, inactivation kinetics and rate of recovery from inactivation in a calcium-dependent and isoform-specific manner (PubMed:11847232, PubMed:18957440, PubMed:23576435). Modulates KCND3/Kv4.3 currents (PubMed:23576435). Isoform 4 does not increase KCND2 expression at the cell membrane (PubMed:18957440). Isoform 4 retains KCND3 in the endoplasmic reticulum and negatively regulates its expression at the cell membrane.
Regulatory subunit of Kv4/D (Shal)-type voltage-gated rapidly inactivating A-type potassium channels. Modulates KCND2 channel density, inactivation kinetics and rate of recovery from inactivation in a calcium-dependent and isoform-specific manner (PubMed:11847232, PubMed:18957440, PubMed:23576435). Modulates KCND3/Kv4.3 currents (PubMed:23576435). Isoform 4 does not increase KCND2 expression at the cell membrane (PubMed:18957440). Isoform 4 retains KCND3 in the endoplasmic reticulum and negatively regulates its expression at the cell membrane.
Subunit
Component of heteromultimeric potassium channels (PubMed:19713751, PubMed:20943905). Identified in potassium channel complexes containing KCND1, KCND2, KCND3, KCNIP1, KCNIP2, KCNIP3, KCNIP4, DPP6 and DPP10 (PubMed:19713751). Interacts with the C-terminus of PSEN2 and probably PSEN1 (By similarity). Interacts with KCND2 and KCND3.
Component of heteromultimeric potassium channels (PubMed:23576435). Identified in potassium channel complexes containing KCND1, KCND2, KCND3, KCNIP1, KCNIP2, KCNIP3, KCNIP4, DPP6 and DPP10 (By similarity). Interacts with KCND2 (PubMed:11847232, PubMed:18957440). Interacts with KCND3 (By similarity). Interacts with the C-terminus of PSEN2 and probably PSEN1 (PubMed:11847232).
Component of heteromultimeric potassium channels (PubMed:23576435). Identified in potassium channel complexes containing KCND1, KCND2, KCND3, KCNIP1, KCNIP2, KCNIP3, KCNIP4, DPP6 and DPP10 (By similarity). Interacts with KCND2 (PubMed:11847232, PubMed:18957440). Interacts with KCND3 (By similarity). Interacts with the C-terminus of PSEN2 and probably PSEN1 (PubMed:11847232).
Similarity
Belongs to the recoverin family.
Keywords
3D-structure
Alternative splicing
Calcium
Cell membrane
Complete proteome
Cytoplasm
Ion channel
Ion transport
Membrane
Metal-binding
Phosphoprotein
Potassium
Potassium channel
Potassium transport
Reference proteome
Repeat
Transport
Voltage-gated channel
Endoplasmic reticulum
Feature
chain Kv channel-interacting protein 4
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
A0A0N1PGQ2
A0A212FJX2
A0A0N1IDK7
A0A1W4XFA3
D6W6Q6
A0A1B6D535
+ More
A0A224XX92 E2B4K6 A0A3L8DLR7 A0A0C9RFF9 A0A0C9QJ24 A0A088AF31 V9IB71 V9I9E5 A0A0J7LBP7 E2A8M0 E9IMW8 K7IV73 A0A158NH56 A0A232FJI0 A0A2S2R4R2 A0A151J4J6 A0A226E7C5 A0A195AZL6 A0A2S2PCK1 A0A2S2PGU9 J9JSJ5 A0A1D2NN66 A0A026X2K0 A0A1S4MXN0 E0VYI0 F4X070 A0A151X5R6 A0A0L7QYG7 A0A0M8ZYR4 E9G3C0 A0A164UUB4 A0A0P6B721 A0A195D1H6 A0A0P6I3T6 A0A151JVY6 A0A0L8GY77 A0A2G8L355 A0A2A3ET84 K1QKC5 A0A210QRQ4 A0A1V9XRA3 A0A0L8GYK1 A0A1S3I2L0 T1JUV0 A0A1E1XND2 A0A1S3I2K8 A0A1V9X612 A0A131XSJ6 A0A087USP3 T1J5U2 F6V8I4 V4A303 Q640X4 Q6R4N5 A0A3P8V8W6 A0A1W3JP88 A0A1W5B4Z1 Q5RDR0 A0A286XA43 A0A3Q3JW50 A0A094MDK1 A0A226PD84 A0A1S3I3U6 K7G7D7 A0A3Q3JZF6 Q5DGE2 A0A2K5W367 Q6PHZ8-3 Q6PIL6-3 A0A0G2KAZ2 Q3V060 A0A0D9RVH6 A0A3Q7RSD0 A0A2Y9G830 A0A337RVK3 A0A2U3WZS4 A0A1D5QKI0 A0A3Q7WZA2 A0A2J8ULA5 A0A1S2ZYC8 A0A2J8NW46 Q7PMH8 A0A1U8D8F1 G5B471 A0A1U7RMT2
A0A224XX92 E2B4K6 A0A3L8DLR7 A0A0C9RFF9 A0A0C9QJ24 A0A088AF31 V9IB71 V9I9E5 A0A0J7LBP7 E2A8M0 E9IMW8 K7IV73 A0A158NH56 A0A232FJI0 A0A2S2R4R2 A0A151J4J6 A0A226E7C5 A0A195AZL6 A0A2S2PCK1 A0A2S2PGU9 J9JSJ5 A0A1D2NN66 A0A026X2K0 A0A1S4MXN0 E0VYI0 F4X070 A0A151X5R6 A0A0L7QYG7 A0A0M8ZYR4 E9G3C0 A0A164UUB4 A0A0P6B721 A0A195D1H6 A0A0P6I3T6 A0A151JVY6 A0A0L8GY77 A0A2G8L355 A0A2A3ET84 K1QKC5 A0A210QRQ4 A0A1V9XRA3 A0A0L8GYK1 A0A1S3I2L0 T1JUV0 A0A1E1XND2 A0A1S3I2K8 A0A1V9X612 A0A131XSJ6 A0A087USP3 T1J5U2 F6V8I4 V4A303 Q640X4 Q6R4N5 A0A3P8V8W6 A0A1W3JP88 A0A1W5B4Z1 Q5RDR0 A0A286XA43 A0A3Q3JW50 A0A094MDK1 A0A226PD84 A0A1S3I3U6 K7G7D7 A0A3Q3JZF6 Q5DGE2 A0A2K5W367 Q6PHZ8-3 Q6PIL6-3 A0A0G2KAZ2 Q3V060 A0A0D9RVH6 A0A3Q7RSD0 A0A2Y9G830 A0A337RVK3 A0A2U3WZS4 A0A1D5QKI0 A0A3Q7WZA2 A0A2J8ULA5 A0A1S2ZYC8 A0A2J8NW46 Q7PMH8 A0A1U8D8F1 G5B471 A0A1U7RMT2
Pubmed
26354079
22118469
18362917
19820115
20798317
30249741
+ More
21282665 20075255 21347285 28648823 27289101 24508170 20566863 21719571 21292972 29023486 22992520 28812685 28327890 29209593 12481130 15114417 23254933 16449567 24487278 21993624 24621616 17381049 16617374 11847232 11805342 16141072 15489334 15363885 19713751 21183079 20943905 20045463 19109250 16112838 14702039 15815621 18957440 23576435 24811166 15057822 15632090 10349636 11042159 11076861 11217851 12466851 12040188 16141073 17975172 17431167 12364791 21993625
21282665 20075255 21347285 28648823 27289101 24508170 20566863 21719571 21292972 29023486 22992520 28812685 28327890 29209593 12481130 15114417 23254933 16449567 24487278 21993624 24621616 17381049 16617374 11847232 11805342 16141072 15489334 15363885 19713751 21183079 20943905 20045463 19109250 16112838 14702039 15815621 18957440 23576435 24811166 15057822 15632090 10349636 11042159 11076861 11217851 12466851 12040188 16141073 17975172 17431167 12364791 21993625
EMBL
KQ460400
KPJ14990.1
AGBW02008207
OWR54009.1
KQ459071
KPJ03860.1
+ More
KQ971307 EFA10959.2 GEDC01016542 JAS20756.1 GFTR01003274 JAW13152.1 GL445574 EFN89359.1 QOIP01000006 RLU21375.1 GBYB01005726 JAG75493.1 GBYB01003554 JAG73321.1 JR037322 AEY57711.1 JR037321 AEY57710.1 LBMM01000017 KMR05324.1 KMR05332.1 GL437652 EFN70207.1 GL764255 EFZ18066.1 AAZX01005118 ADTU01015586 ADTU01015587 NNAY01000129 OXU30700.1 GGMS01015751 MBY84954.1 KQ980126 KYN17561.1 LNIX01000006 OXA52791.1 KQ976698 KYM77482.1 GGMR01014495 MBY27114.1 GGMR01015517 MBY28136.1 ABLF02038998 LJIJ01000003 ODN06525.1 KK107020 EZA62512.1 AAZO01006258 AAZO01006259 AAZO01006260 DS235845 EEB18436.1 GL888493 EGI60135.1 KQ982510 KYQ55630.1 KQ414688 KOC63634.1 KQ435821 KOX72343.1 GL732531 EFX86013.1 LRGB01001574 KZS11675.1 GDIP01019274 JAM84441.1 KQ976973 KYN06722.1 GDIQ01010871 JAN83866.1 KQ981682 KYN38076.1 KQ419933 KOF81923.1 MRZV01000238 PIK54683.1 KZ288185 PBC34907.1 JH818480 EKC37182.1 NEDP02002268 OWF51401.1 MNPL01005415 OQR76036.1 KOF81924.1 CAEY01000784 GFAA01002597 JAU00838.1 MNPL01022450 OQR69035.1 GEFM01006169 JAP69627.1 KK121387 KFM80382.1 JH431868 EAAA01002224 KB202284 ESO91082.1 BC082465 AAH82465.1 AY514488 AAS00647.1 CR857842 CAH90097.1 AAKN02022804 AAKN02022805 AAKN02022806 KL336331 KFZ50116.1 AWGT02000101 OXB77419.1 AGCU01099107 AGCU01099108 AGCU01099109 AGCU01099110 AGCU01099111 AGCU01099112 AGCU01099113 AGCU01099114 AGCU01099115 AGCU01099116 AY813382 AAW25114.1 AQIA01053958 AQIA01053959 AQIA01053960 AQIA01053961 AQIA01053962 AQIA01053963 AQIA01053964 AQIA01053965 AQIA01053966 AQIA01053967 AF305071 AF453243 AY647240 AK039048 BC051130 CK618709 AF302044 AF305072 AF453246 AF367023 AF367024 AY029176 AY118170 DQ148487 DQ148488 DQ148489 DQ148490 DQ148491 AK289922 AC096576 AC097505 AC098597 AC104065 AC107462 AC108147 AC109360 AC109636 AC110296 AC110612 AC113606 AC116641 CH471069 BC032520 AABR07015610 AABR07015611 AABR07015612 CH473963 EDL99910.1 AK133416 CH466524 BAE21645.1 EDL37638.1 AQIB01114551 AQIB01114552 AANG04001605 JSUE03033025 JSUE03033026 JSUE03033027 JSUE03033028 JSUE03033029 JSUE03033030 JSUE03033031 JSUE03033032 JSUE03033033 JSUE03033034 NDHI03003454 PNJ46049.1 PNJ46051.1 NBAG03000222 PNI75981.1 PNI75983.1 AAAB01008980 EAA13817.5 JH168398 EHB04082.1
KQ971307 EFA10959.2 GEDC01016542 JAS20756.1 GFTR01003274 JAW13152.1 GL445574 EFN89359.1 QOIP01000006 RLU21375.1 GBYB01005726 JAG75493.1 GBYB01003554 JAG73321.1 JR037322 AEY57711.1 JR037321 AEY57710.1 LBMM01000017 KMR05324.1 KMR05332.1 GL437652 EFN70207.1 GL764255 EFZ18066.1 AAZX01005118 ADTU01015586 ADTU01015587 NNAY01000129 OXU30700.1 GGMS01015751 MBY84954.1 KQ980126 KYN17561.1 LNIX01000006 OXA52791.1 KQ976698 KYM77482.1 GGMR01014495 MBY27114.1 GGMR01015517 MBY28136.1 ABLF02038998 LJIJ01000003 ODN06525.1 KK107020 EZA62512.1 AAZO01006258 AAZO01006259 AAZO01006260 DS235845 EEB18436.1 GL888493 EGI60135.1 KQ982510 KYQ55630.1 KQ414688 KOC63634.1 KQ435821 KOX72343.1 GL732531 EFX86013.1 LRGB01001574 KZS11675.1 GDIP01019274 JAM84441.1 KQ976973 KYN06722.1 GDIQ01010871 JAN83866.1 KQ981682 KYN38076.1 KQ419933 KOF81923.1 MRZV01000238 PIK54683.1 KZ288185 PBC34907.1 JH818480 EKC37182.1 NEDP02002268 OWF51401.1 MNPL01005415 OQR76036.1 KOF81924.1 CAEY01000784 GFAA01002597 JAU00838.1 MNPL01022450 OQR69035.1 GEFM01006169 JAP69627.1 KK121387 KFM80382.1 JH431868 EAAA01002224 KB202284 ESO91082.1 BC082465 AAH82465.1 AY514488 AAS00647.1 CR857842 CAH90097.1 AAKN02022804 AAKN02022805 AAKN02022806 KL336331 KFZ50116.1 AWGT02000101 OXB77419.1 AGCU01099107 AGCU01099108 AGCU01099109 AGCU01099110 AGCU01099111 AGCU01099112 AGCU01099113 AGCU01099114 AGCU01099115 AGCU01099116 AY813382 AAW25114.1 AQIA01053958 AQIA01053959 AQIA01053960 AQIA01053961 AQIA01053962 AQIA01053963 AQIA01053964 AQIA01053965 AQIA01053966 AQIA01053967 AF305071 AF453243 AY647240 AK039048 BC051130 CK618709 AF302044 AF305072 AF453246 AF367023 AF367024 AY029176 AY118170 DQ148487 DQ148488 DQ148489 DQ148490 DQ148491 AK289922 AC096576 AC097505 AC098597 AC104065 AC107462 AC108147 AC109360 AC109636 AC110296 AC110612 AC113606 AC116641 CH471069 BC032520 AABR07015610 AABR07015611 AABR07015612 CH473963 EDL99910.1 AK133416 CH466524 BAE21645.1 EDL37638.1 AQIB01114551 AQIB01114552 AANG04001605 JSUE03033025 JSUE03033026 JSUE03033027 JSUE03033028 JSUE03033029 JSUE03033030 JSUE03033031 JSUE03033032 JSUE03033033 JSUE03033034 NDHI03003454 PNJ46049.1 PNJ46051.1 NBAG03000222 PNI75981.1 PNI75983.1 AAAB01008980 EAA13817.5 JH168398 EHB04082.1
Proteomes
UP000053240
UP000007151
UP000053268
UP000192223
UP000007266
UP000008237
+ More
UP000279307 UP000005203 UP000036403 UP000000311 UP000002358 UP000005205 UP000215335 UP000078492 UP000198287 UP000078540 UP000007819 UP000094527 UP000053097 UP000009046 UP000007755 UP000075809 UP000053825 UP000053105 UP000000305 UP000076858 UP000078542 UP000078541 UP000053454 UP000230750 UP000242457 UP000005408 UP000242188 UP000192247 UP000085678 UP000015104 UP000054359 UP000008144 UP000030746 UP000265120 UP000005447 UP000261600 UP000198419 UP000007267 UP000233100 UP000000589 UP000005640 UP000002494 UP000029965 UP000286641 UP000248481 UP000011712 UP000245340 UP000006718 UP000286642 UP000079721 UP000007062 UP000189705 UP000006813
UP000279307 UP000005203 UP000036403 UP000000311 UP000002358 UP000005205 UP000215335 UP000078492 UP000198287 UP000078540 UP000007819 UP000094527 UP000053097 UP000009046 UP000007755 UP000075809 UP000053825 UP000053105 UP000000305 UP000076858 UP000078542 UP000078541 UP000053454 UP000230750 UP000242457 UP000005408 UP000242188 UP000192247 UP000085678 UP000015104 UP000054359 UP000008144 UP000030746 UP000265120 UP000005447 UP000261600 UP000198419 UP000007267 UP000233100 UP000000589 UP000005640 UP000002494 UP000029965 UP000286641 UP000248481 UP000011712 UP000245340 UP000006718 UP000286642 UP000079721 UP000007062 UP000189705 UP000006813
Pfam
Interpro
SUPFAM
SSF47473
SSF47473
CDD
ProteinModelPortal
A0A0N1PGQ2
A0A212FJX2
A0A0N1IDK7
A0A1W4XFA3
D6W6Q6
A0A1B6D535
+ More
A0A224XX92 E2B4K6 A0A3L8DLR7 A0A0C9RFF9 A0A0C9QJ24 A0A088AF31 V9IB71 V9I9E5 A0A0J7LBP7 E2A8M0 E9IMW8 K7IV73 A0A158NH56 A0A232FJI0 A0A2S2R4R2 A0A151J4J6 A0A226E7C5 A0A195AZL6 A0A2S2PCK1 A0A2S2PGU9 J9JSJ5 A0A1D2NN66 A0A026X2K0 A0A1S4MXN0 E0VYI0 F4X070 A0A151X5R6 A0A0L7QYG7 A0A0M8ZYR4 E9G3C0 A0A164UUB4 A0A0P6B721 A0A195D1H6 A0A0P6I3T6 A0A151JVY6 A0A0L8GY77 A0A2G8L355 A0A2A3ET84 K1QKC5 A0A210QRQ4 A0A1V9XRA3 A0A0L8GYK1 A0A1S3I2L0 T1JUV0 A0A1E1XND2 A0A1S3I2K8 A0A1V9X612 A0A131XSJ6 A0A087USP3 T1J5U2 F6V8I4 V4A303 Q640X4 Q6R4N5 A0A3P8V8W6 A0A1W3JP88 A0A1W5B4Z1 Q5RDR0 A0A286XA43 A0A3Q3JW50 A0A094MDK1 A0A226PD84 A0A1S3I3U6 K7G7D7 A0A3Q3JZF6 Q5DGE2 A0A2K5W367 Q6PHZ8-3 Q6PIL6-3 A0A0G2KAZ2 Q3V060 A0A0D9RVH6 A0A3Q7RSD0 A0A2Y9G830 A0A337RVK3 A0A2U3WZS4 A0A1D5QKI0 A0A3Q7WZA2 A0A2J8ULA5 A0A1S2ZYC8 A0A2J8NW46 Q7PMH8 A0A1U8D8F1 G5B471 A0A1U7RMT2
A0A224XX92 E2B4K6 A0A3L8DLR7 A0A0C9RFF9 A0A0C9QJ24 A0A088AF31 V9IB71 V9I9E5 A0A0J7LBP7 E2A8M0 E9IMW8 K7IV73 A0A158NH56 A0A232FJI0 A0A2S2R4R2 A0A151J4J6 A0A226E7C5 A0A195AZL6 A0A2S2PCK1 A0A2S2PGU9 J9JSJ5 A0A1D2NN66 A0A026X2K0 A0A1S4MXN0 E0VYI0 F4X070 A0A151X5R6 A0A0L7QYG7 A0A0M8ZYR4 E9G3C0 A0A164UUB4 A0A0P6B721 A0A195D1H6 A0A0P6I3T6 A0A151JVY6 A0A0L8GY77 A0A2G8L355 A0A2A3ET84 K1QKC5 A0A210QRQ4 A0A1V9XRA3 A0A0L8GYK1 A0A1S3I2L0 T1JUV0 A0A1E1XND2 A0A1S3I2K8 A0A1V9X612 A0A131XSJ6 A0A087USP3 T1J5U2 F6V8I4 V4A303 Q640X4 Q6R4N5 A0A3P8V8W6 A0A1W3JP88 A0A1W5B4Z1 Q5RDR0 A0A286XA43 A0A3Q3JW50 A0A094MDK1 A0A226PD84 A0A1S3I3U6 K7G7D7 A0A3Q3JZF6 Q5DGE2 A0A2K5W367 Q6PHZ8-3 Q6PIL6-3 A0A0G2KAZ2 Q3V060 A0A0D9RVH6 A0A3Q7RSD0 A0A2Y9G830 A0A337RVK3 A0A2U3WZS4 A0A1D5QKI0 A0A3Q7WZA2 A0A2J8ULA5 A0A1S2ZYC8 A0A2J8NW46 Q7PMH8 A0A1U8D8F1 G5B471 A0A1U7RMT2
PDB
3DD4
E-value=1.83874e-44,
Score=448
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
Cytoplasm
Endoplasmic reticulum
Cytoplasm
Endoplasmic reticulum
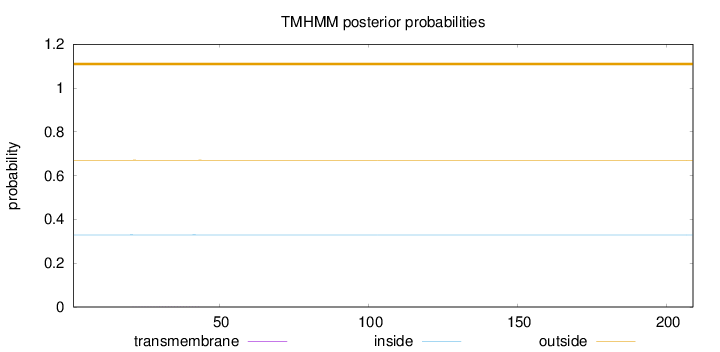
Length:
209
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01349
Exp number, first 60 AAs:
0.01219
Total prob of N-in:
0.32924
outside
1 - 209
Population Genetic Test Statistics
Pi
236.00814
Theta
160.92243
Tajima's D
1.631422
CLR
0.292229
CSRT
0.813859307034648
Interpretation
Uncertain
