Pre Gene Modal
BGIBMGA011987
Annotation
PREDICTED:_RNA_pseudouridylate_synthase_domain-containing_protein_1-like_isoform_X3_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.106 Nuclear Reliability : 1.692
Sequence
CDS
ATGGCTGTGGAAATTCTATTTGAAAACTCTGATTATTTAATCGTGAACAAGCCTTATGACATGTATATCAATTCTGATGACGAAAACGAAAAGAACACAGTGGCTTACCAAATAGCGCGTTGCGATTCTCATCTGTCAACGTCATCGCAACCTTTACATTTCGTCCAACGATTGGACTACGCCACGAGTGGCGTGCTCTGCATGGCCAAGAGTCGGCCAGCTGCGGCCAGCGCTGGCAAAGCGTTCGAAAAGAGATTGACCAGGAAATACTACCTGGCGATCGTCCGCGGACATGTTGGCTTTGAGCTGGCCGATGTTGATTATGGCGTTGGCGCAGACCCTGAAACCCTGCACTCGAATCACAAGATGCTGGCGCGAACATCTTCATCTCAAACGATACAAGTTCGCAGCGCTCAGACCAGGATGCTGCTCTTGGAGACGGGCTGGTACCAACGGAGTCCTGTGTCTGTACTCCTACTGAAGCCTATAACTGGTCGTCGCCATCAGCTGAGGGTCCACTGTAACGCAATAGGCCACACTATACTGGGCGATTACACTTACAGCGACAGGTCGGACTGTTCTCCACACCGCATGTTTCTCCACGCTTCCAGGCTAGTTTTACCAGTCGACAAGGAAGTTATCGACGTTCAAACCAAAACCCCTTACTTTTCCGATGAAGGCTTCCTCCAGCACTGGAAGTCTGATCGGGTAGCGTACCCTTACAAAACCAAAGGCGACTTCAATGATGTATGTGACGTGATCGATAAGTCATATGTGCTGGGCGTCAACTATAAAATGTATAATTGCACCAATTAA
Protein
MAVEILFENSDYLIVNKPYDMYINSDDENEKNTVAYQIARCDSHLSTSSQPLHFVQRLDYATSGVLCMAKSRPAAASAGKAFEKRLTRKYYLAIVRGHVGFELADVDYGVGADPETLHSNHKMLARTSSSQTIQVRSAQTRMLLLETGWYQRSPVSVLLLKPITGRRHQLRVHCNAIGHTILGDYTYSDRSDCSPHRMFLHASRLVLPVDKEVIDVQTKTPYFSDEGFLQHWKSDRVAYPYKTKGDFNDVCDVIDKSYVLGVNYKMYNCTN
Summary
Uniprot
H9JR27
A0A3S2LS15
A0A2A4K5H1
A0A194Q264
A0A212FH41
A0A2H1V5J7
+ More
A0A2Z5U7A8 A0A2W1BCP9 A0A1B6E762 E9HPA1 A0A067QYS9 A0A1B6JPT6 A0A1B6G9A7 A0A0P6E1Y5 A0A0P5NER4 A0A0P5J4E2 A0A0N8AVL8 A0A0P5RHR0 A0A0P6ELN9 A0A2R7VWS0 A0A0P6HBX8 A0A2J7R5A5 A0A1B6L096 A0A0P5E194 T1JGR8 A0A0P5XUN6 A0A164RTJ5 A0A0P5WYQ3 A0A0P6B974 E2CAE1 A0A232F8C0 A0A023F456 A0A154PU29 A0A0L7RIS3 Q171X7 A0A182H0K9 K7IPA2 A0A1Q3G2M1 A0A1Q3G2K2 W5JBZ9 A0A2M4CSY2 A0A0N0BIR2 A0A182NCU1 A0A182QSJ8 A0A182PLG2 A0A182SWU8 A0A182JC68 A0A182UTE8 A0A182RI33 A0A182W376 A0A026X0M3 A0A182KZT6 A0A182XF59 A0A182HVN0 A0A151XCA4 A0A226EKL8 A0A182MUS0 A0A2G8KGU7 C3ZJW0 A0A2S2NLP3 A0A195F1A7 F4X6M2 A0A2H8TVG4 A0A3R7M658 A0A182JZ78 Q7PQ70 A0A195B2K9 A0A182YQE8 A0A158NLC6 E9IZ73 A0A195E7H1 A0A1S3IXX3 A0A0C9RL97 D6W746 A0A146KMY7 A0A146LKN6 A0A0A9XYW7 A0A146LRY7 A0A0P5T7Y6 A0A0K8S7L7 A0A0P5TG43 A0A2P6K0P7 V5H7G3 B7P2L1 A0A182FAE6 E2AT95 T1FYI1 A0A1S3DBI3 A0A087UID3 R7UFN6 A0A2A3EA45 N6UKF4 A0A091ERN0 A0A0L8IDN0
A0A2Z5U7A8 A0A2W1BCP9 A0A1B6E762 E9HPA1 A0A067QYS9 A0A1B6JPT6 A0A1B6G9A7 A0A0P6E1Y5 A0A0P5NER4 A0A0P5J4E2 A0A0N8AVL8 A0A0P5RHR0 A0A0P6ELN9 A0A2R7VWS0 A0A0P6HBX8 A0A2J7R5A5 A0A1B6L096 A0A0P5E194 T1JGR8 A0A0P5XUN6 A0A164RTJ5 A0A0P5WYQ3 A0A0P6B974 E2CAE1 A0A232F8C0 A0A023F456 A0A154PU29 A0A0L7RIS3 Q171X7 A0A182H0K9 K7IPA2 A0A1Q3G2M1 A0A1Q3G2K2 W5JBZ9 A0A2M4CSY2 A0A0N0BIR2 A0A182NCU1 A0A182QSJ8 A0A182PLG2 A0A182SWU8 A0A182JC68 A0A182UTE8 A0A182RI33 A0A182W376 A0A026X0M3 A0A182KZT6 A0A182XF59 A0A182HVN0 A0A151XCA4 A0A226EKL8 A0A182MUS0 A0A2G8KGU7 C3ZJW0 A0A2S2NLP3 A0A195F1A7 F4X6M2 A0A2H8TVG4 A0A3R7M658 A0A182JZ78 Q7PQ70 A0A195B2K9 A0A182YQE8 A0A158NLC6 E9IZ73 A0A195E7H1 A0A1S3IXX3 A0A0C9RL97 D6W746 A0A146KMY7 A0A146LKN6 A0A0A9XYW7 A0A146LRY7 A0A0P5T7Y6 A0A0K8S7L7 A0A0P5TG43 A0A2P6K0P7 V5H7G3 B7P2L1 A0A182FAE6 E2AT95 T1FYI1 A0A1S3DBI3 A0A087UID3 R7UFN6 A0A2A3EA45 N6UKF4 A0A091ERN0 A0A0L8IDN0
Pubmed
19121390
26354079
22118469
28756777
21292972
24845553
+ More
20798317 28648823 25474469 17510324 26483478 20075255 20920257 23761445 24508170 30249741 20966253 29023486 18563158 21719571 12364791 14747013 17210077 25244985 21347285 21282665 18362917 19820115 26823975 25401762 25765539 23254933 23537049
20798317 28648823 25474469 17510324 26483478 20075255 20920257 23761445 24508170 30249741 20966253 29023486 18563158 21719571 12364791 14747013 17210077 25244985 21347285 21282665 18362917 19820115 26823975 25401762 25765539 23254933 23537049
EMBL
BABH01028682
BABH01028683
RSAL01000312
RVE42661.1
NWSH01000138
PCG79168.1
+ More
KQ459564 KPI99631.1 AGBW02008548 OWR53052.1 ODYU01000772 SOQ36069.1 AP017511 BBB06829.1 KZ150211 PZC72191.1 GEDC01003527 JAS33771.1 GL732704 EFX66443.1 KK852818 KDR15682.1 GECU01006438 JAT01269.1 GECZ01010744 JAS59025.1 GDIQ01069376 JAN25361.1 GDIQ01165840 JAK85885.1 GDIQ01207216 JAK44509.1 GDIQ01238515 JAK13210.1 GDIQ01119797 JAL31929.1 GDIQ01067967 JAN26770.1 KK854135 PTY11912.1 GDIQ01022368 JAN72369.1 NEVH01007392 PNF36000.1 GEBQ01023457 GEBQ01023053 JAT16520.1 JAT16924.1 GDIP01149346 JAJ74056.1 JH432211 GDIP01067068 JAM36647.1 LRGB01002140 KZS08928.1 GDIP01079660 JAM24055.1 GDIP01018510 JAM85205.1 GL453975 EFN75107.1 NNAY01000743 OXU26730.1 GBBI01002933 JAC15779.1 KQ435119 KZC14710.1 KQ414583 KOC70734.1 CH477443 EAT40809.1 JXUM01101465 JXUM01101466 KQ564651 KXJ71955.1 GFDL01001056 JAV33989.1 GFDL01001015 JAV34030.1 ADMH02001950 ETN60359.1 GGFL01004259 MBW68437.1 KQ435727 KOX77922.1 AXCN02001206 KK107063 QOIP01000005 EZA60949.1 RLU22510.1 APCN01005806 KQ982314 KYQ58011.1 LNIX01000003 OXA57291.1 AXCM01000387 MRZV01000594 PIK47216.1 GG666633 EEN47245.1 GGMR01005484 MBY18103.1 KQ981864 KYN34355.1 GL888818 EGI57804.1 GFXV01006449 MBW18254.1 QCYY01001950 ROT74030.1 AAAB01008898 EAA09234.6 KQ976662 KYM78439.1 ADTU01019320 GL767119 EFZ14121.1 KQ979568 KYN20784.1 GBYB01008129 GBYB01014112 JAG77896.1 JAG83879.1 KQ971307 EFA11504.2 GDHC01021783 JAP96845.1 GDHC01010101 JAQ08528.1 GBHO01021114 JAG22490.1 GDHC01009107 JAQ09522.1 GDIP01136156 JAL67558.1 GBRD01016683 JAG49144.1 GDIP01128108 JAL75606.1 MWRG01049905 PRD19904.1 GANP01005418 JAB79050.1 ABJB010144409 DS622764 EEC00833.1 GL442545 EFN63340.1 AMQM01000987 KB096742 ESO02682.1 KK119940 KFM77122.1 AMQN01009004 KB304376 ELU02087.1 KZ288310 PBC28597.1 APGK01020355 KB740193 KB631669 ENN81116.1 ERL85230.1 KK718885 KFO58959.1 KQ415935 KOF99582.1
KQ459564 KPI99631.1 AGBW02008548 OWR53052.1 ODYU01000772 SOQ36069.1 AP017511 BBB06829.1 KZ150211 PZC72191.1 GEDC01003527 JAS33771.1 GL732704 EFX66443.1 KK852818 KDR15682.1 GECU01006438 JAT01269.1 GECZ01010744 JAS59025.1 GDIQ01069376 JAN25361.1 GDIQ01165840 JAK85885.1 GDIQ01207216 JAK44509.1 GDIQ01238515 JAK13210.1 GDIQ01119797 JAL31929.1 GDIQ01067967 JAN26770.1 KK854135 PTY11912.1 GDIQ01022368 JAN72369.1 NEVH01007392 PNF36000.1 GEBQ01023457 GEBQ01023053 JAT16520.1 JAT16924.1 GDIP01149346 JAJ74056.1 JH432211 GDIP01067068 JAM36647.1 LRGB01002140 KZS08928.1 GDIP01079660 JAM24055.1 GDIP01018510 JAM85205.1 GL453975 EFN75107.1 NNAY01000743 OXU26730.1 GBBI01002933 JAC15779.1 KQ435119 KZC14710.1 KQ414583 KOC70734.1 CH477443 EAT40809.1 JXUM01101465 JXUM01101466 KQ564651 KXJ71955.1 GFDL01001056 JAV33989.1 GFDL01001015 JAV34030.1 ADMH02001950 ETN60359.1 GGFL01004259 MBW68437.1 KQ435727 KOX77922.1 AXCN02001206 KK107063 QOIP01000005 EZA60949.1 RLU22510.1 APCN01005806 KQ982314 KYQ58011.1 LNIX01000003 OXA57291.1 AXCM01000387 MRZV01000594 PIK47216.1 GG666633 EEN47245.1 GGMR01005484 MBY18103.1 KQ981864 KYN34355.1 GL888818 EGI57804.1 GFXV01006449 MBW18254.1 QCYY01001950 ROT74030.1 AAAB01008898 EAA09234.6 KQ976662 KYM78439.1 ADTU01019320 GL767119 EFZ14121.1 KQ979568 KYN20784.1 GBYB01008129 GBYB01014112 JAG77896.1 JAG83879.1 KQ971307 EFA11504.2 GDHC01021783 JAP96845.1 GDHC01010101 JAQ08528.1 GBHO01021114 JAG22490.1 GDHC01009107 JAQ09522.1 GDIP01136156 JAL67558.1 GBRD01016683 JAG49144.1 GDIP01128108 JAL75606.1 MWRG01049905 PRD19904.1 GANP01005418 JAB79050.1 ABJB010144409 DS622764 EEC00833.1 GL442545 EFN63340.1 AMQM01000987 KB096742 ESO02682.1 KK119940 KFM77122.1 AMQN01009004 KB304376 ELU02087.1 KZ288310 PBC28597.1 APGK01020355 KB740193 KB631669 ENN81116.1 ERL85230.1 KK718885 KFO58959.1 KQ415935 KOF99582.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000000305
+ More
UP000027135 UP000235965 UP000076858 UP000008237 UP000215335 UP000076502 UP000053825 UP000008820 UP000069940 UP000249989 UP000002358 UP000000673 UP000053105 UP000075884 UP000075886 UP000075885 UP000075901 UP000075880 UP000075903 UP000075900 UP000075920 UP000053097 UP000279307 UP000075882 UP000076407 UP000075840 UP000075809 UP000198287 UP000075883 UP000230750 UP000001554 UP000078541 UP000007755 UP000283509 UP000075881 UP000007062 UP000078540 UP000076408 UP000005205 UP000078492 UP000085678 UP000007266 UP000001555 UP000069272 UP000000311 UP000015101 UP000079169 UP000054359 UP000014760 UP000242457 UP000019118 UP000030742 UP000052976 UP000053454
UP000027135 UP000235965 UP000076858 UP000008237 UP000215335 UP000076502 UP000053825 UP000008820 UP000069940 UP000249989 UP000002358 UP000000673 UP000053105 UP000075884 UP000075886 UP000075885 UP000075901 UP000075880 UP000075903 UP000075900 UP000075920 UP000053097 UP000279307 UP000075882 UP000076407 UP000075840 UP000075809 UP000198287 UP000075883 UP000230750 UP000001554 UP000078541 UP000007755 UP000283509 UP000075881 UP000007062 UP000078540 UP000076408 UP000005205 UP000078492 UP000085678 UP000007266 UP000001555 UP000069272 UP000000311 UP000015101 UP000079169 UP000054359 UP000014760 UP000242457 UP000019118 UP000030742 UP000052976 UP000053454
PRIDE
Pfam
PF00849 PseudoU_synth_2
Interpro
ProteinModelPortal
H9JR27
A0A3S2LS15
A0A2A4K5H1
A0A194Q264
A0A212FH41
A0A2H1V5J7
+ More
A0A2Z5U7A8 A0A2W1BCP9 A0A1B6E762 E9HPA1 A0A067QYS9 A0A1B6JPT6 A0A1B6G9A7 A0A0P6E1Y5 A0A0P5NER4 A0A0P5J4E2 A0A0N8AVL8 A0A0P5RHR0 A0A0P6ELN9 A0A2R7VWS0 A0A0P6HBX8 A0A2J7R5A5 A0A1B6L096 A0A0P5E194 T1JGR8 A0A0P5XUN6 A0A164RTJ5 A0A0P5WYQ3 A0A0P6B974 E2CAE1 A0A232F8C0 A0A023F456 A0A154PU29 A0A0L7RIS3 Q171X7 A0A182H0K9 K7IPA2 A0A1Q3G2M1 A0A1Q3G2K2 W5JBZ9 A0A2M4CSY2 A0A0N0BIR2 A0A182NCU1 A0A182QSJ8 A0A182PLG2 A0A182SWU8 A0A182JC68 A0A182UTE8 A0A182RI33 A0A182W376 A0A026X0M3 A0A182KZT6 A0A182XF59 A0A182HVN0 A0A151XCA4 A0A226EKL8 A0A182MUS0 A0A2G8KGU7 C3ZJW0 A0A2S2NLP3 A0A195F1A7 F4X6M2 A0A2H8TVG4 A0A3R7M658 A0A182JZ78 Q7PQ70 A0A195B2K9 A0A182YQE8 A0A158NLC6 E9IZ73 A0A195E7H1 A0A1S3IXX3 A0A0C9RL97 D6W746 A0A146KMY7 A0A146LKN6 A0A0A9XYW7 A0A146LRY7 A0A0P5T7Y6 A0A0K8S7L7 A0A0P5TG43 A0A2P6K0P7 V5H7G3 B7P2L1 A0A182FAE6 E2AT95 T1FYI1 A0A1S3DBI3 A0A087UID3 R7UFN6 A0A2A3EA45 N6UKF4 A0A091ERN0 A0A0L8IDN0
A0A2Z5U7A8 A0A2W1BCP9 A0A1B6E762 E9HPA1 A0A067QYS9 A0A1B6JPT6 A0A1B6G9A7 A0A0P6E1Y5 A0A0P5NER4 A0A0P5J4E2 A0A0N8AVL8 A0A0P5RHR0 A0A0P6ELN9 A0A2R7VWS0 A0A0P6HBX8 A0A2J7R5A5 A0A1B6L096 A0A0P5E194 T1JGR8 A0A0P5XUN6 A0A164RTJ5 A0A0P5WYQ3 A0A0P6B974 E2CAE1 A0A232F8C0 A0A023F456 A0A154PU29 A0A0L7RIS3 Q171X7 A0A182H0K9 K7IPA2 A0A1Q3G2M1 A0A1Q3G2K2 W5JBZ9 A0A2M4CSY2 A0A0N0BIR2 A0A182NCU1 A0A182QSJ8 A0A182PLG2 A0A182SWU8 A0A182JC68 A0A182UTE8 A0A182RI33 A0A182W376 A0A026X0M3 A0A182KZT6 A0A182XF59 A0A182HVN0 A0A151XCA4 A0A226EKL8 A0A182MUS0 A0A2G8KGU7 C3ZJW0 A0A2S2NLP3 A0A195F1A7 F4X6M2 A0A2H8TVG4 A0A3R7M658 A0A182JZ78 Q7PQ70 A0A195B2K9 A0A182YQE8 A0A158NLC6 E9IZ73 A0A195E7H1 A0A1S3IXX3 A0A0C9RL97 D6W746 A0A146KMY7 A0A146LKN6 A0A0A9XYW7 A0A146LRY7 A0A0P5T7Y6 A0A0K8S7L7 A0A0P5TG43 A0A2P6K0P7 V5H7G3 B7P2L1 A0A182FAE6 E2AT95 T1FYI1 A0A1S3DBI3 A0A087UID3 R7UFN6 A0A2A3EA45 N6UKF4 A0A091ERN0 A0A0L8IDN0
PDB
5VBB
E-value=1.82836e-34,
Score=363
Ontologies
KEGG
GO
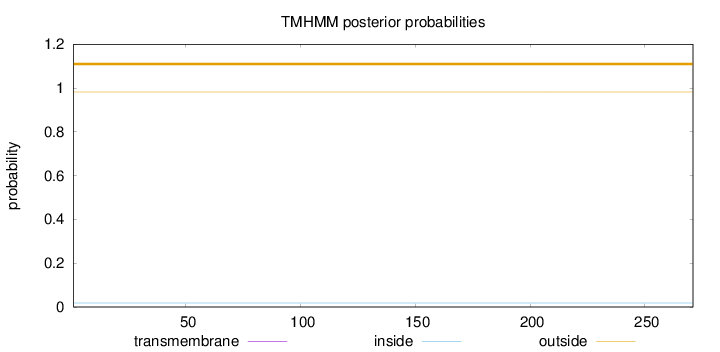
Topology
Length:
271
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00198
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.01777
outside
1 - 271
Population Genetic Test Statistics
Pi
234.373678
Theta
185.849848
Tajima's D
0.133839
CLR
0.429227
CSRT
0.407929603519824
Interpretation
Uncertain
