Gene
KWMTBOMO06334 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011901
Annotation
sericin_2_isoform_1_precursor_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.504
Sequence
CDS
ATGAAGATCCCATACGTCTTGCTGTTCCTTGTGGGCGTGGCTGTGGTCAACGCATTGCCCAATCCACTTTTCGGGGGCTTAGTTAAATCGCTTAGCAAAAAAAAACAAATTTTTGAGGACAAATTTGAAAATCTCAAAGAGAATGTGGGTGAAAAATTTGAAAATCTCAAAGAAAACGTGGGTGAAAAAGTTGAAAATCTCAAAGAGAATGTTGGTGAAAAATTAGAAAATATCAAAGAGAAGGCTGGAGAAAAATTTGAAAATCTCAAAGACAATGTTGGAGAAAAATTTGAAAATCTCAAAGACAATGTTGGAGACAAATTAGAAGCAGCTAAAGAAAAAGCTGGAGAAATCAAAAAGAAACTAGTGGATGTCGGCGAAGATCTGAAAGACGAGCTCACGGAAGACAAAAAAATAAAGATATCCATCTCGAAAGACGAAGGATTGACTTTAGAAAAAGAAGGATACAAGTCTGATTACGATCGTAATGAATATGAAGAACGCGGATCTGAACACCAGGAGGATAACGACAGCGACGGCTCATACAGCAAAGGTTCCGAATACGAAAAATACGGTGAAGAGGAAAAGTATGAAGAACGTAGGACCCACGACAAGTTCTCCATTGGCAAGAACGGTATTAGTGCCGAGCGAACGAAATCAAAGAGAGGTGAAAGAAAAGAAGTCGAAGGCGAATATGAGAAAGACTACGAAAGGAAAGAGAACAACGGCGGATCCTCCGAATATTCAGAGAGAGAAAGAGAGAGTTTGGAAAAATCAAAAGAAAGGTACGGCGAGCAGTCGTCTAAGTCCTTCTCGTTGGGTAAATCCGGCTTGAAGAAGCAGGATAATTCAAAATCGTATTCCGACAAGGAAGAGTCGAAACTGGAAAAGGAAAAGAAATATGAAAAGAAAACAAAAATCAACAATGAAAGGCAGTTAGATGAAGATGAGAACGAAAGGAGAACTGTTGTAGGCCGAGATGAGCAAAGGCAAGATGACCAAAGCCGAGACGACCAAAGCCGAGATGACCAAAGCCAAGATGAGGAAACTGGCAGCGACGATAGTGACAAAAATAGAGGAAAGGATACTGACGATAAATATTCCGAGACAGGAACCAATAAATCATCAGAAACGAAGACAGGCAAGCGTGATGGCTCGAAGAGCGGCGTCACAGTCGAAAGGGAAAAATCCGAATCCAACAAGAAAAGTCGTGAATTTGAAAATAAAGAAGCTGAATCTTCGACCTATAGGGATAAGAATCGGTCAGTGAACAGTGGCTCGGAACGCAAGAGTTCCGGTAAAGACGAGGAGTACAGTGAACAGAACTCCAGTAATAAATCCTTTAACGACGGCGATGCATCGGCTGACTACCAAACCAAATCTAAGAAGGTTGAAAAGAATTCTGCCAGAGATAAAAAGGAAAAGGAAAAAACTGACACAAGAAATTCTGACGGAACGTACAAAACTTCTGAGCGCGAAAAAGAACAATCTTCTAGAGTGAATCAAAGTAAGGGCAGCAACTCTCGGGATTCCTCGGAGTCAGACAAATCTGGCCGAAAAGTGAATAAAGAAACAGAAACGTACTCTGACAAAGACGCGCAGACTTCAGAAAGTGAACGTACTCAAAGTAAAGAAAAAAAAAATACAGCGCCCAAGAATAAGGGCAAAAAGGGAACCTCTACAGAAACAGATGGAGTCACTAAGAATGCTAGTAAGCAGAAGGAGAAGGTGCCTAAAGATGGAAGTAAAAGCTCCACGAATGATAGCGAAGGCAAACAAAAAAACAAAGACCAATCAAAGGGACAGAAAAATAATCAAGACGGACAAGACTCTTCGACGAACGAAAATTCCAAAAAGACAGATGATAATGTTGCAAAGAAAGAAGAACCCAATAATCAAAAGAGAGAACAAAAAGGAAAGACAAGATGTGGCTCTAGGAAAACTGAGAGTTCCAAAGCGAAAGAAGATAGAAGCAAGAAAAGCACTACCGATAAGGACCAGCGCGACGACAAAAAAGATTCCTCCAGCAAAAACATAGACAAGCCTAAAGATGGGTCATCATCGGATAAAGACTCAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGTAAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGTAAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGTAAAGCCTAACGACAGATCACCAACGCATAAAGACACAGAGAAGGTAAAACCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGTAAAGCCTAACGACAGATCACCATCGTATAAAGACACTGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGCTAAGCATAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGTAAAACCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGTATAAAGACACTGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGCTAAGCATAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGTAAAACCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGTAAAGCCTAACGACAGATCACAATCGTATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAAGCTAAGCCTAACGGCAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGTAAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAGAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAAGCTAAGCCTAACGGCAGATCACCATCGGATAAAGACACAGAGAAGGTAAAGCCTAACGACAGATCACCATCGCATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAAGCTAAGCCTAACGGCAGATCACCATCGGATAAAGACACAGAGAAGGTAAAGCCTAACGACAGATCACCATCGCATAAATACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCACCATCGGATAAAGACACAGATAAGACATTCGACAAAAACATAGACAATAAAAGGCCCAAGGACGGTTCATCATCGGATAAGAACGTAGAGCAGGAAAGGGAGAACTACAAATCCGAATCTAGCCGAAATGAATTTGAAAACCAAAAATCTGCACATTCGAGATATGAGGATAACGGTGGTTTGAAAGAAAAAAGCTCGCAATCTAAGAATTACGGAAGAGACGAGAAGTACAGTGAAGAGAAGGAGAGGAGTTCCACTGGGAAGTTTGGATCGAATGACTCCCGAGCACGTTCTACAAAAGCCGAAGAGGAACATGTCAGGAAGTCACAAGAAGAAACACATTCAGAGCAACGAGAAAAAACAAGATCTGACGGAGTGACTAAATACAACGATGGTGATGAGCATTTCGATTCAGACGATACAGAGAAGACTAAGCCTAATGGCAGATCACCATCGCATAAAGACACAGAGAAGGCTAAGCCTAACGACAGATCATCCTCGGATAAAGACACAGAGAAGACATTTGACAAAAACATAGACAATAAAAGGCCCAAGGACGGTTCATCATCGGATAAGAACGTAGAGCAGGAAAGGGAGAACTACAAATCCGAATCTAGCCGAAATGAATTTGAAAACCAAAAATCTGCACATTCGAGATATGAGGATAACGGTGGTTTGAAAGAAAAAAGCTCGCAATCTAAGAATTACGGAAGAGACGAGAAGTACAGTGAAGAGAAGGAGAGGAGTTCCACTGGGAAGTCTGGATCGAATGACTCCCGAGCACGTTCTACAAAAGCCGAAGAGGAACATGTCAGGAAGTCACAAGAAGAAACACATTCAGAGCAACGAGGAAGAACAAGATCTGACGGAGCAACTACGTCCAACGATAATGATAAGCAATACGATTCAGACGACAAGAACAACAGTAGCACCAAACATAAGAAAACGGTCATGAGATCTGAGCAATCAGATTCGTCTCAAAATGAAAATTCTACCTCCGAATCCAAAAAGTTCGCAAAAACTGATGGCTCCAACAAATATGAAGCGGAATCAAGTTCGCATAAACAACAAGAAGCGCGCAAACAGAGCAATAGAGTAGTAGAGAAGAGCACTGATGGGGACAATGAGGAATCCTACAGGAGCGAAAGCAGCAGCAGCAGCAGCAGCAGTTCTAGTAGTAGTCGAAGCAGTTCCTCAAGCACCTACACCGGCTCCCATGATGATAGCTCCGAGGAATGA
Protein
MKIPYVLLFLVGVAVVNALPNPLFGGLVKSLSKKKQIFEDKFENLKENVGEKFENLKENVGEKVENLKENVGEKLENIKEKAGEKFENLKDNVGEKFENLKDNVGDKLEAAKEKAGEIKKKLVDVGEDLKDELTEDKKIKISISKDEGLTLEKEGYKSDYDRNEYEERGSEHQEDNDSDGSYSKGSEYEKYGEEEKYEERRTHDKFSIGKNGISAERTKSKRGERKEVEGEYEKDYERKENNGGSSEYSERERESLEKSKERYGEQSSKSFSLGKSGLKKQDNSKSYSDKEESKLEKEKKYEKKTKINNERQLDEDENERRTVVGRDEQRQDDQSRDDQSRDDQSQDEETGSDDSDKNRGKDTDDKYSETGTNKSSETKTGKRDGSKSGVTVEREKSESNKKSREFENKEAESSTYRDKNRSVNSGSERKSSGKDEEYSEQNSSNKSFNDGDASADYQTKSKKVEKNSARDKKEKEKTDTRNSDGTYKTSEREKEQSSRVNQSKGSNSRDSSESDKSGRKVNKETETYSDKDAQTSESERTQSKEKKNTAPKNKGKKGTSTETDGVTKNASKQKEKVPKDGSKSSTNDSEGKQKNKDQSKGQKNNQDGQDSSTNENSKKTDDNVAKKEEPNNQKREQKGKTRCGSRKTESSKAKEDRSKKSTTDKDQRDDKKDSSSKNIDKPKDGSSSDKDSEKAKPNDRSPSHKDTEKVKPNDRSPSHKDTEKVKPNDRSPSDKDTEKAKPNDRSPSHKDTEKVKPNDRSPTHKDTEKVKPNDRSPSHKDTEKAKPNDRSPSDKDTEKAKPNDRSPSHKDTEKVKPNDRSPSYKDTEKAKPNDRSPSDKDTEKAKPNDRSPSHKDTEKAKHNDRSPSDKDTEKAKPNDRSPSHKDTEKAKPNDRSPSHKDTEKVKPNDRSPSHKDTEKAKPNDRSPSDKDTEKAKPNDRSPSYKDTEKAKPNDRSPSDKDTEKAKPNDRSPSHKDTEKAKHNDRSPSDKDTEKAKPNDRSPSHKDTEKAKPNDRSPSHKDTEKVKPNDRSPSHKDTEKAKPNDRSPSDKDTEKAKPNDRSPSHKDTEKVKPNDRSQSYKDTEKAKPNDRSPSDKDTEKAKPNGRSPSDKDTEKAKPNDRSPSHKDTEKVKPNDRSPSHKDTEKAKPNDRSPSDRDTEKAKPNDRSPSDKDTEKAKPNGRSPSDKDTEKVKPNDRSPSHKDTEKAKPNDRSPSDKDTEKAKPNDRSPSDKDTEKAKPNDRSPSDKDTEKAKPNDRSPSDKDTEKAKPNGRSPSDKDTEKVKPNDRSPSHKYTEKAKPNDRSPSDKDTEKAKPNDRSPSDKDTEKAKPNDRSPSDKDTDKTFDKNIDNKRPKDGSSSDKNVEQERENYKSESSRNEFENQKSAHSRYEDNGGLKEKSSQSKNYGRDEKYSEEKERSSTGKFGSNDSRARSTKAEEEHVRKSQEETHSEQREKTRSDGVTKYNDGDEHFDSDDTEKTKPNGRSPSHKDTEKAKPNDRSSSDKDTEKTFDKNIDNKRPKDGSSSDKNVEQERENYKSESSRNEFENQKSAHSRYEDNGGLKEKSSQSKNYGRDEKYSEEKERSSTGKSGSNDSRARSTKAEEEHVRKSQEETHSEQRGRTRSDGATTSNDNDKQYDSDDKNNSSTKHKKTVMRSEQSDSSQNENSTSESKKFAKTDGSNKYEAESSSHKQQEARKQSNRVVEKSTDGDNEESYRSESSSSSSSSSSSSRSSSSSTYTGSHDDSSEE
Summary
Uniprot
Pubmed
EMBL
PRIDE
ProteinModelPortal
PDB
5JCS
E-value=0.00715257,
Score=99
Ontologies
GO
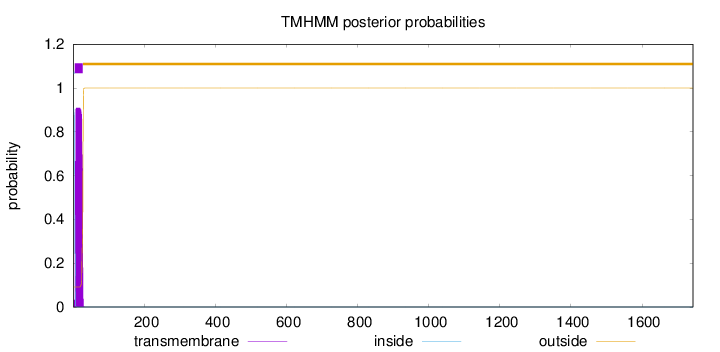
Topology
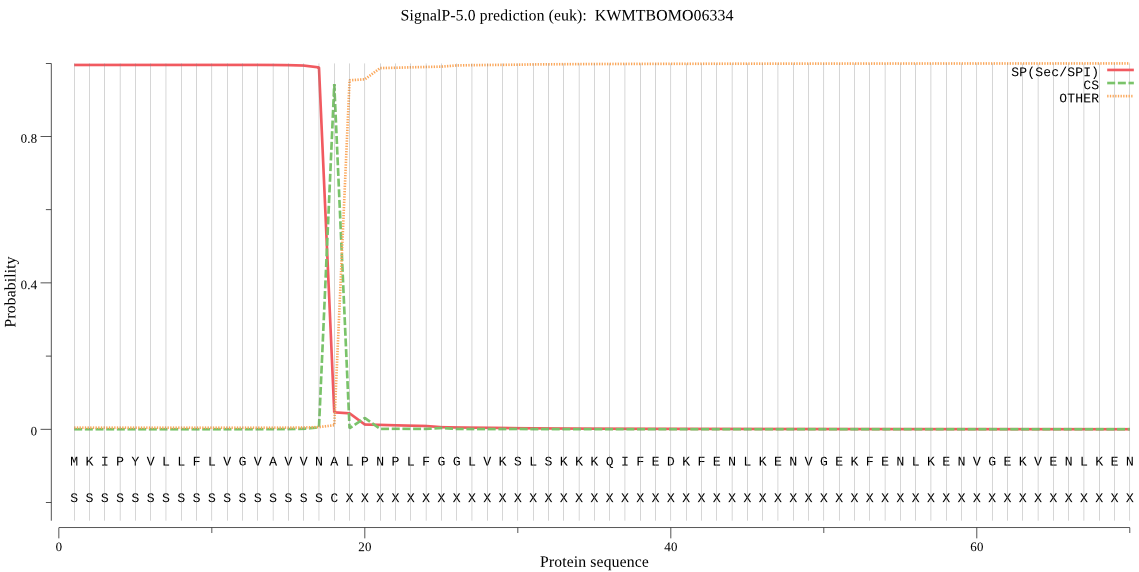
SignalP
Position: 1 - 18,
Likelihood: 0.995181
Length:
1743
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.00258
Exp number, first 60 AAs:
20.00258
Total prob of N-in:
0.90920
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 1743
Population Genetic Test Statistics
Pi
209.078537
Theta
182.909976
Tajima's D
-0.863054
CLR
0.623853
CSRT
0.166591670416479
Interpretation
Uncertain
