Pre Gene Modal
BGIBMGA011898
Annotation
Motile_sperm_domain-containing_protein_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 2.387
Sequence
CDS
ATGGCTACTGCGGCGGACGTGCGGAGCATTTTTGAGAATCGTCTCAAAGAAAACGATGTAGACTTCCACCCGGCCGACTTAGAGCGAACGAGAGATGAGAAGTACCTGAAGAGGGTGCTGGCCCATTGCGAGAAAGACGCTAAGCAGGCTGCCGACATGCTGTGGGACATTCTTACCTGGAGGAAAAATGTCGGAGCTTCCAACATCAACGAGGACAACATACGCATGGACTATGTTAAAGAAGGTGTCTTCTTCCCGCGGGGCAGAGACGTGGATGGCAGTCTCCTCCTCGTCTTCAAATCCAAAATGCACATCAAAGGCCAAAGGGACTATGACGAGTTGAAAAAAATCATGATATACTGGTTCGATAGGGTCGAGCGAGAAGAGAATGGTAAAAAAATCAGCATGTTCTTTGATATGGACGGTACTGGCCTCAGTAACATGGACATGGAACTAATCAAATACTTAATAACCCTATTTAAATGTTATTATCCGTATTTCCTCAACTACATCATTATTTACAACATGCCCTGGGTTTTGTCGGCGGCTTTTAAAGTGGTGAAGACATTGTTACCTGCCAAGGCCGTTGAAAAAATGAAATTTGTCACCAAAGATACGTTGAAGGATATTGTGGCACCGGAACAAGCTCTTGCCTGTTGGGGAGGTAAAGACAACTATGTTTTTGAATTCATACCAGAAAGTCGTAGTTCAGAGCATACTCCGAAGAAAGTCACATTTGCAGAGCAGAGCGAAAATCAGCACTCGCTCGGCGAGATGCTCAGAATTGTTCCGAATGATACTATAATATTCAAAACAGACAATGATGAGATCTCCGGGCAGTTCACCATAACAAATATGGATGAGAGTGCGATATCATTCAAAATTCGTACAACTTCACCAGAAAAGTTCCGGGTTAGACCCAGCTCGGGTACTCTAGCAGCTGGGATGTCACAATCAGTCTTAATAGTTGTCCAACCTGGTTTTCATCTACGAACAGTTACAAAGGATCGCTTTCTGGTAATGTCAGTACAAATACCAAAGACAGATATGACATCAAAAGATCTAGCGGACATATGGCAGAGTTCGCCTAGTAGTAAAATAGATGAATACCGTTTAAAATGCCAATTCCCGGAGAAGGATTTGCCTAAAAATGGCAATTTGCTTGACAAAGTCTCCGAGAAAGACTCACTGTCAAATTCATTGAATACCCTGCAGGTCCATTATGAGTTATTAAACAGAGACGTTAAAAGAATGAAAATATTTCAGTTTCTGACACTGCTATTCTCCGGGGTGTCGGTGGTACTTGGCTACCTCATTTACAACTCCACAAGCATCGAAGACAGGTATTGCGAAAGAATGTAG
Protein
MATAADVRSIFENRLKENDVDFHPADLERTRDEKYLKRVLAHCEKDAKQAADMLWDILTWRKNVGASNINEDNIRMDYVKEGVFFPRGRDVDGSLLLVFKSKMHIKGQRDYDELKKIMIYWFDRVEREENGKKISMFFDMDGTGLSNMDMELIKYLITLFKCYYPYFLNYIIIYNMPWVLSAAFKVVKTLLPAKAVEKMKFVTKDTLKDIVAPEQALACWGGKDNYVFEFIPESRSSEHTPKKVTFAEQSENQHSLGEMLRIVPNDTIIFKTDNDEISGQFTITNMDESAISFKIRTTSPEKFRVRPSSGTLAAGMSQSVLIVVQPGFHLRTVTKDRFLVMSVQIPKTDMTSKDLADIWQSSPSSKIDEYRLKCQFPEKDLPKNGNLLDKVSEKDSLSNSLNTLQVHYELLNRDVKRMKIFQFLTLLFSGVSVVLGYLIYNSTSIEDRYCERM
Summary
Uniprot
H9JQU2
A0A2W1BQG5
A0A2A4JX08
A0A2H1WZP6
A0A0L7L2G7
A0A0N1PI63
+ More
A0A1E1W359 A0A194Q878 A0A212FH33 A0A2J7RP83 A0A2P8Y0S1 A0A067RJ94 A0A1S4MXW5 D6WGY2 A0A1Y1MZQ2 A0A026WE23 E2C1R1 A0A195CTF8 A0A195BJK1 A0A158NFC8 E0VZQ7 F4WUJ4 A0A151JZJ8 A0A151XFN8 A0A195E9M2 A0A1B6LNH2 A0A146MDY3 A0A1B6CK24 A0A0A9YF94 R4G3Q1 A0A232ENM4 A0A0A9YF99 A0A3Q0IZL9 A0A0P4VHS8 N6TYX8 A0A1L8DYX9 E2A6C4 U4U6G3 A0A023F886 A0A224XF82 A0A2S2PCT6 A0A0T6BG97 A0A084WKL3 A0A0L7R8I5 A0A182IJT8 A0A2A3E587 J9K0C1 A0A154NZ19 A0A088A5E4 K7J6A9 A0A182VG93 A0A2H8TF15 A0A2C9GPH8 A0A182LJE0 Q7Q717 A0A182PW97 E9HJL8 A0A182RMG7 A0A0J7KZB1 A0A2R7W1H0 A0A182LVN4 A0A182Y126 A0A1Q3G2D9 A0A182WKE6 A0A1Q3G299 Q16GR0 A0A0P6BVI5 A0A1I8MH83 A0A2M4BLD5 A0A162TC47 A0A0P5YZR7 A0A0P5LIB0 A0A0N8EB75 A0A0P6HFH0 W5JXC8 A0A1I8QFA8 A0A0P6H0D6 A0A1I8QF52 A0A1I8QF51 A0A0L0CSF5 W8AQ77 A0A0P5ZKG3 A0A1B0CKR3 U5EYC4 A0A0P5RIE5 A0A0P6D5S2 A0A0P5LLI2 A0A0P6FAS4 A0A0P5Y9P9 A0A0P4Y0F7 A0A182FCH9 A0A0P6CPU9 A0A034VIZ3 A0A1W4UX70 B4HUL4 A0A1B0FK55 A0A0J9RQ07
A0A1E1W359 A0A194Q878 A0A212FH33 A0A2J7RP83 A0A2P8Y0S1 A0A067RJ94 A0A1S4MXW5 D6WGY2 A0A1Y1MZQ2 A0A026WE23 E2C1R1 A0A195CTF8 A0A195BJK1 A0A158NFC8 E0VZQ7 F4WUJ4 A0A151JZJ8 A0A151XFN8 A0A195E9M2 A0A1B6LNH2 A0A146MDY3 A0A1B6CK24 A0A0A9YF94 R4G3Q1 A0A232ENM4 A0A0A9YF99 A0A3Q0IZL9 A0A0P4VHS8 N6TYX8 A0A1L8DYX9 E2A6C4 U4U6G3 A0A023F886 A0A224XF82 A0A2S2PCT6 A0A0T6BG97 A0A084WKL3 A0A0L7R8I5 A0A182IJT8 A0A2A3E587 J9K0C1 A0A154NZ19 A0A088A5E4 K7J6A9 A0A182VG93 A0A2H8TF15 A0A2C9GPH8 A0A182LJE0 Q7Q717 A0A182PW97 E9HJL8 A0A182RMG7 A0A0J7KZB1 A0A2R7W1H0 A0A182LVN4 A0A182Y126 A0A1Q3G2D9 A0A182WKE6 A0A1Q3G299 Q16GR0 A0A0P6BVI5 A0A1I8MH83 A0A2M4BLD5 A0A162TC47 A0A0P5YZR7 A0A0P5LIB0 A0A0N8EB75 A0A0P6HFH0 W5JXC8 A0A1I8QFA8 A0A0P6H0D6 A0A1I8QF52 A0A1I8QF51 A0A0L0CSF5 W8AQ77 A0A0P5ZKG3 A0A1B0CKR3 U5EYC4 A0A0P5RIE5 A0A0P6D5S2 A0A0P5LLI2 A0A0P6FAS4 A0A0P5Y9P9 A0A0P4Y0F7 A0A182FCH9 A0A0P6CPU9 A0A034VIZ3 A0A1W4UX70 B4HUL4 A0A1B0FK55 A0A0J9RQ07
Pubmed
19121390
28756777
26227816
26354079
22118469
29403074
+ More
24845553 20566863 18362917 19820115 28004739 24508170 30249741 20798317 21347285 21719571 26823975 25401762 28648823 27129103 23537049 25474469 24438588 20075255 20966253 12364791 14747013 17210077 21292972 25244985 17510324 25315136 20920257 23761445 26108605 24495485 25348373 17994087 22936249
24845553 20566863 18362917 19820115 28004739 24508170 30249741 20798317 21347285 21719571 26823975 25401762 28648823 27129103 23537049 25474469 24438588 20075255 20966253 12364791 14747013 17210077 21292972 25244985 17510324 25315136 20920257 23761445 26108605 24495485 25348373 17994087 22936249
EMBL
BABH01028707
KZ150077
PZC73943.1
NWSH01000452
PCG76316.1
ODYU01012288
+ More
SOQ58521.1 JTDY01003483 KOB69484.1 KQ459736 KPJ20224.1 GDQN01009660 JAT81394.1 KQ459564 KPI99610.1 AGBW02008548 OWR53051.1 NEVH01002140 PNF42647.1 PYGN01001081 PSN37774.1 KK852434 KDR23946.1 AAZO01006526 KQ971321 EFA00139.1 GEZM01016826 JAV91134.1 KK107260 QOIP01000001 EZA54168.1 RLU26621.1 GL451975 EFN78120.1 KQ977279 KYN03983.1 KQ976464 KYM84544.1 ADTU01014261 ADTU01014262 DS235854 EEB18863.1 GL888365 EGI62159.1 KQ981410 KYN41923.1 KQ982182 KYQ59189.1 KQ979440 KYN21507.1 GEBQ01014695 JAT25282.1 GDHC01001582 JAQ17047.1 GEDC01023536 JAS13762.1 GBHO01013821 JAG29783.1 ACPB03014131 GAHY01001466 JAA76044.1 NNAY01003104 OXU19969.1 GBHO01013816 JAG29788.1 GDKW01002394 JAI54201.1 APGK01049569 KB741156 ENN73551.1 GFDF01002411 JAV11673.1 GL437123 EFN71009.1 KB632169 ERL89469.1 GBBI01001176 JAC17536.1 GFTR01006742 JAW09684.1 GGMR01014389 MBY27008.1 LJIG01000588 KRT86352.1 ATLV01024126 KE525349 KFB50757.1 KQ414633 KOC67076.1 KZ288364 PBC26897.1 ABLF02038391 KQ434783 KZC04919.1 GFXV01000860 GFXV01002280 MBW12665.1 MBW14085.1 APCN01004270 APCN01004271 AAAB01008960 EAA11055.4 GL732663 EFX68040.1 LBMM01001814 KMQ95701.1 KK854202 PTY13031.1 AXCM01000864 GFDL01001081 JAV33964.1 GFDL01001100 JAV33945.1 CH478244 EAT33429.1 GDIP01016922 JAM86793.1 GGFJ01004735 MBW53876.1 LRGB01000005 KZS22072.1 GDIP01051161 JAM52554.1 GDIQ01180068 JAK71657.1 GDIQ01043868 JAN50869.1 GDIQ01020179 JAN74558.1 ADMH02000008 ETN68114.1 GDIQ01036639 JAN58098.1 JRES01000080 KNC34334.1 GAMC01019920 GAMC01019919 GAMC01019917 JAB86638.1 GDIP01042398 JAM61317.1 AJWK01016546 AJWK01016547 AJWK01016548 AJWK01016549 GANO01000450 JAB59421.1 GDIQ01101157 JAL50569.1 GDIQ01090498 JAN04239.1 GDIQ01178614 JAK73111.1 GDIQ01052024 JAN42713.1 GDIP01062410 JAM41305.1 GDIP01234340 JAI89061.1 GDIP01011181 JAM92534.1 GAKP01017424 JAC41528.1 CH480817 EDW50635.1 CCAG010003333 CCAG010003334 CM002912 KMY97827.1
SOQ58521.1 JTDY01003483 KOB69484.1 KQ459736 KPJ20224.1 GDQN01009660 JAT81394.1 KQ459564 KPI99610.1 AGBW02008548 OWR53051.1 NEVH01002140 PNF42647.1 PYGN01001081 PSN37774.1 KK852434 KDR23946.1 AAZO01006526 KQ971321 EFA00139.1 GEZM01016826 JAV91134.1 KK107260 QOIP01000001 EZA54168.1 RLU26621.1 GL451975 EFN78120.1 KQ977279 KYN03983.1 KQ976464 KYM84544.1 ADTU01014261 ADTU01014262 DS235854 EEB18863.1 GL888365 EGI62159.1 KQ981410 KYN41923.1 KQ982182 KYQ59189.1 KQ979440 KYN21507.1 GEBQ01014695 JAT25282.1 GDHC01001582 JAQ17047.1 GEDC01023536 JAS13762.1 GBHO01013821 JAG29783.1 ACPB03014131 GAHY01001466 JAA76044.1 NNAY01003104 OXU19969.1 GBHO01013816 JAG29788.1 GDKW01002394 JAI54201.1 APGK01049569 KB741156 ENN73551.1 GFDF01002411 JAV11673.1 GL437123 EFN71009.1 KB632169 ERL89469.1 GBBI01001176 JAC17536.1 GFTR01006742 JAW09684.1 GGMR01014389 MBY27008.1 LJIG01000588 KRT86352.1 ATLV01024126 KE525349 KFB50757.1 KQ414633 KOC67076.1 KZ288364 PBC26897.1 ABLF02038391 KQ434783 KZC04919.1 GFXV01000860 GFXV01002280 MBW12665.1 MBW14085.1 APCN01004270 APCN01004271 AAAB01008960 EAA11055.4 GL732663 EFX68040.1 LBMM01001814 KMQ95701.1 KK854202 PTY13031.1 AXCM01000864 GFDL01001081 JAV33964.1 GFDL01001100 JAV33945.1 CH478244 EAT33429.1 GDIP01016922 JAM86793.1 GGFJ01004735 MBW53876.1 LRGB01000005 KZS22072.1 GDIP01051161 JAM52554.1 GDIQ01180068 JAK71657.1 GDIQ01043868 JAN50869.1 GDIQ01020179 JAN74558.1 ADMH02000008 ETN68114.1 GDIQ01036639 JAN58098.1 JRES01000080 KNC34334.1 GAMC01019920 GAMC01019919 GAMC01019917 JAB86638.1 GDIP01042398 JAM61317.1 AJWK01016546 AJWK01016547 AJWK01016548 AJWK01016549 GANO01000450 JAB59421.1 GDIQ01101157 JAL50569.1 GDIQ01090498 JAN04239.1 GDIQ01178614 JAK73111.1 GDIQ01052024 JAN42713.1 GDIP01062410 JAM41305.1 GDIP01234340 JAI89061.1 GDIP01011181 JAM92534.1 GAKP01017424 JAC41528.1 CH480817 EDW50635.1 CCAG010003333 CCAG010003334 CM002912 KMY97827.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000235965 UP000245037 UP000027135 UP000007266 UP000053097 UP000279307 UP000008237 UP000078542 UP000078540 UP000005205 UP000009046 UP000007755 UP000078541 UP000075809 UP000078492 UP000015103 UP000215335 UP000079169 UP000019118 UP000000311 UP000030742 UP000030765 UP000053825 UP000075880 UP000242457 UP000007819 UP000076502 UP000005203 UP000002358 UP000075903 UP000075840 UP000075882 UP000007062 UP000075885 UP000000305 UP000075900 UP000036403 UP000075883 UP000076408 UP000075920 UP000008820 UP000095301 UP000076858 UP000000673 UP000095300 UP000037069 UP000092461 UP000069272 UP000192221 UP000001292 UP000092444
UP000235965 UP000245037 UP000027135 UP000007266 UP000053097 UP000279307 UP000008237 UP000078542 UP000078540 UP000005205 UP000009046 UP000007755 UP000078541 UP000075809 UP000078492 UP000015103 UP000215335 UP000079169 UP000019118 UP000000311 UP000030742 UP000030765 UP000053825 UP000075880 UP000242457 UP000007819 UP000076502 UP000005203 UP000002358 UP000075903 UP000075840 UP000075882 UP000007062 UP000075885 UP000000305 UP000075900 UP000036403 UP000075883 UP000076408 UP000075920 UP000008820 UP000095301 UP000076858 UP000000673 UP000095300 UP000037069 UP000092461 UP000069272 UP000192221 UP000001292 UP000092444
PRIDE
Pfam
Interpro
IPR036865
CRAL-TRIO_dom_sf
+ More
IPR036273 CRAL/TRIO_N_dom_sf
IPR008962 PapD-like_sf
IPR013783 Ig-like_fold
IPR001251 CRAL-TRIO_dom
IPR000535 MSP_dom
IPR017453 GCV_H_sub
IPR002930 GCV_H
IPR033753 GCV_H/Fam206
IPR003016 2-oxoA_DH_lipoyl-BS
IPR000089 Biotin_lipoyl
IPR011053 Single_hybrid_motif
IPR027902 DUF4487
IPR042231 Cho/carn_acyl_trans_2
IPR042232 Cho/carn_acyl_trans_1
IPR039551 Cho/carn_acyl_trans
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR032476 CPT_N
IPR000542 Carn_acyl_trans
IPR036273 CRAL/TRIO_N_dom_sf
IPR008962 PapD-like_sf
IPR013783 Ig-like_fold
IPR001251 CRAL-TRIO_dom
IPR000535 MSP_dom
IPR017453 GCV_H_sub
IPR002930 GCV_H
IPR033753 GCV_H/Fam206
IPR003016 2-oxoA_DH_lipoyl-BS
IPR000089 Biotin_lipoyl
IPR011053 Single_hybrid_motif
IPR027902 DUF4487
IPR042231 Cho/carn_acyl_trans_2
IPR042232 Cho/carn_acyl_trans_1
IPR039551 Cho/carn_acyl_trans
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR032476 CPT_N
IPR000542 Carn_acyl_trans
SUPFAM
Gene 3D
ProteinModelPortal
H9JQU2
A0A2W1BQG5
A0A2A4JX08
A0A2H1WZP6
A0A0L7L2G7
A0A0N1PI63
+ More
A0A1E1W359 A0A194Q878 A0A212FH33 A0A2J7RP83 A0A2P8Y0S1 A0A067RJ94 A0A1S4MXW5 D6WGY2 A0A1Y1MZQ2 A0A026WE23 E2C1R1 A0A195CTF8 A0A195BJK1 A0A158NFC8 E0VZQ7 F4WUJ4 A0A151JZJ8 A0A151XFN8 A0A195E9M2 A0A1B6LNH2 A0A146MDY3 A0A1B6CK24 A0A0A9YF94 R4G3Q1 A0A232ENM4 A0A0A9YF99 A0A3Q0IZL9 A0A0P4VHS8 N6TYX8 A0A1L8DYX9 E2A6C4 U4U6G3 A0A023F886 A0A224XF82 A0A2S2PCT6 A0A0T6BG97 A0A084WKL3 A0A0L7R8I5 A0A182IJT8 A0A2A3E587 J9K0C1 A0A154NZ19 A0A088A5E4 K7J6A9 A0A182VG93 A0A2H8TF15 A0A2C9GPH8 A0A182LJE0 Q7Q717 A0A182PW97 E9HJL8 A0A182RMG7 A0A0J7KZB1 A0A2R7W1H0 A0A182LVN4 A0A182Y126 A0A1Q3G2D9 A0A182WKE6 A0A1Q3G299 Q16GR0 A0A0P6BVI5 A0A1I8MH83 A0A2M4BLD5 A0A162TC47 A0A0P5YZR7 A0A0P5LIB0 A0A0N8EB75 A0A0P6HFH0 W5JXC8 A0A1I8QFA8 A0A0P6H0D6 A0A1I8QF52 A0A1I8QF51 A0A0L0CSF5 W8AQ77 A0A0P5ZKG3 A0A1B0CKR3 U5EYC4 A0A0P5RIE5 A0A0P6D5S2 A0A0P5LLI2 A0A0P6FAS4 A0A0P5Y9P9 A0A0P4Y0F7 A0A182FCH9 A0A0P6CPU9 A0A034VIZ3 A0A1W4UX70 B4HUL4 A0A1B0FK55 A0A0J9RQ07
A0A1E1W359 A0A194Q878 A0A212FH33 A0A2J7RP83 A0A2P8Y0S1 A0A067RJ94 A0A1S4MXW5 D6WGY2 A0A1Y1MZQ2 A0A026WE23 E2C1R1 A0A195CTF8 A0A195BJK1 A0A158NFC8 E0VZQ7 F4WUJ4 A0A151JZJ8 A0A151XFN8 A0A195E9M2 A0A1B6LNH2 A0A146MDY3 A0A1B6CK24 A0A0A9YF94 R4G3Q1 A0A232ENM4 A0A0A9YF99 A0A3Q0IZL9 A0A0P4VHS8 N6TYX8 A0A1L8DYX9 E2A6C4 U4U6G3 A0A023F886 A0A224XF82 A0A2S2PCT6 A0A0T6BG97 A0A084WKL3 A0A0L7R8I5 A0A182IJT8 A0A2A3E587 J9K0C1 A0A154NZ19 A0A088A5E4 K7J6A9 A0A182VG93 A0A2H8TF15 A0A2C9GPH8 A0A182LJE0 Q7Q717 A0A182PW97 E9HJL8 A0A182RMG7 A0A0J7KZB1 A0A2R7W1H0 A0A182LVN4 A0A182Y126 A0A1Q3G2D9 A0A182WKE6 A0A1Q3G299 Q16GR0 A0A0P6BVI5 A0A1I8MH83 A0A2M4BLD5 A0A162TC47 A0A0P5YZR7 A0A0P5LIB0 A0A0N8EB75 A0A0P6HFH0 W5JXC8 A0A1I8QFA8 A0A0P6H0D6 A0A1I8QF52 A0A1I8QF51 A0A0L0CSF5 W8AQ77 A0A0P5ZKG3 A0A1B0CKR3 U5EYC4 A0A0P5RIE5 A0A0P6D5S2 A0A0P5LLI2 A0A0P6FAS4 A0A0P5Y9P9 A0A0P4Y0F7 A0A182FCH9 A0A0P6CPU9 A0A034VIZ3 A0A1W4UX70 B4HUL4 A0A1B0FK55 A0A0J9RQ07
PDB
1WIC
E-value=7.1315e-16,
Score=205
Ontologies
PANTHER
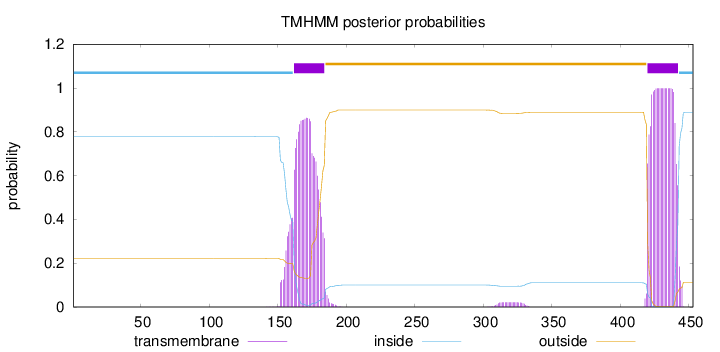
Topology
Length:
453
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
41.10208
Exp number, first 60 AAs:
0
Total prob of N-in:
0.77812
inside
1 - 161
TMhelix
162 - 184
outside
185 - 419
TMhelix
420 - 442
inside
443 - 453
Population Genetic Test Statistics
Pi
224.521559
Theta
198.136993
Tajima's D
0.201263
CLR
0.443569
CSRT
0.428428578571071
Interpretation
Uncertain
