Gene
KWMTBOMO06330
Pre Gene Modal
BGIBMGA011993
Annotation
PREDICTED:_uncharacterized_protein_LOC101740446_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.512 Nuclear Reliability : 1.251
Sequence
CDS
ATGTGTAACGTTATCGATGGCCTGGAAGGGGTCAGCCTGAAGGAAGACTTGTATACGTCAGGGGAATTCAACACGGGGGATTTTGACGAGGATTGGGACGAGCCCGCGGCTGAGGACTCCGATTCTGATTACGAGGATTCGGCCGATGAGGTATACGGGAACCTGCCCCCCGAGCCCAGGTTCGGGAACGTGGAGTACAAGCTGCAGCTGGTGTCGCTCTGCGAGAAACGATTCCAGCATCTGGTCACCCAGCTGAAATGGCGTCTCCGGTCGGGGGGCGGCCGGTGTGTGTACGTGGTGGGAGTGCGGGACTGCGGCGCCGTGGTGGGGCTCCGGGCGCGCGCGCTGAGGGGCTCCCTGCGCACCCTGCGCCGCATGGCGCGCGCCCTCGCCGCCACCCTGCGCGTCACCACCAGGCGTCTGGCGCCGGATCGCGCCGTGGCCGAGGTGCACATACTGAAGCTGGCCGACCCGCAGCAGAGCATGGAGCTGAGGATCGCCGTGATGGGCGCCAACGAAGCTGGGAAGTCCACCTTGATCGGAGTCCTCACGCAAGGCGAGCTGGACAACGGCCGGGGGAGCGCCCGTCTGAACATGTTCCGGCACCTGCACGAGGTCAAGAGCGGCCGGACCTCGTCGCTCAGCCACGAGATACTGGGGTTCGACGCCCAGGGCAACGTGGTGAACTACGGGTGCTCGGAGCTGATGACGGCCGAGCGGATCGGCGAGCGGAGCACCCGGCTGGTGTCGTTCATCGACCTGGCCGGCCACTCCAAGTACCAGCGCACCACGCTGCACGGGCTGGCCGGCTACTCGCCGCACTTCGTCATGCTCGTGATATCGGCGACGGCCGGCATCACGCGCATCACGGAGGAGCACATAGACCTGCTGCTGGCGCTGGACGTGCCGTTCTTCACGGTGGTGACCAAGTGCGAGCTGGCCGCGGCCGCCGTGCAGGACGTCGCCGCCAAGCTGGCCAAGCTCCTGCAGCCCTACAACAAGAGACCACTGCTAATAACAGACGAGAACTCTGCGAGGAACTGCGTGGCCCCACAGAAGCTAATTCTTGACACGATCAATGACAATGCCTCTGAGGATAAATGCGTGATGCCGGAGGTGGCGGTGTTCCCGGTGAGCTGCGTGCGCGGCGCCGGGCTCGGGCCGCTGCACGCCTGCCTCGCCTCGCTCGCTTGTCTACTGGCCACACCCTCCGCCGCGCACTACAACACGCTGCCGCACGACGACACGTGCGAGTTTCAAATCGACGAGATATTCCACGCGGGGGAGTCGTCCGGTCCCGTGGTTGGGGGGCTGCTGGCCCGCGGCAGGATCCGGGAGGGGGACGACCTCCTGATGGGTCCGCTGGACGACGGGCGCTTCGTGTCGGTCCGCGTGGTGGGCGTGCACCGCAACCGGGCCGCCGTGAGCGCCGCGCGCGCCGGACACACGTGCTCGCTGCGACTGCGGGCCCAGCAGGCGCCGCGCCCCGGCATGGTGCTGCTCCGGCAGCCTTCAGACTCTTCGGGCCTTCACTGCAAGAGCAACAACGTGGGGAGCTCTGTGGAGGACGCGATGACCCTGGCCCGAGCTCAGCAAACGCTGTCGAAGCCCGACGCCGACGCGACCAAGAACACGTTCGAAGGAGCCACCGATGACATCCAAGACGACGGCACGACTAATCCTAGGGGATGTCTCTATTTTCAGGCCTCTATACACCTTCTGAGACATTCGACCGCCATCTATAGAGGGTTCCAGTGCACAGTTCACGTCGGCAACGTCACGCAAACAGCCATCATCCAGGGCATCATGTCGGGTACGGGAGAGCTGAGGGCTGGCGAGTCGGCCTCCGCCCTGTTCCGGTTCGTGCGCAGCCCCGAGTACCTGGTGCGCGGCAGGAGGATGTTGTTCACTGCAGGACTCGGCACCAGAGCCATAGGGAGGATAACGCAAGTGTTTCCGCACGTCCCTTGA
Protein
MCNVIDGLEGVSLKEDLYTSGEFNTGDFDEDWDEPAAEDSDSDYEDSADEVYGNLPPEPRFGNVEYKLQLVSLCEKRFQHLVTQLKWRLRSGGGRCVYVVGVRDCGAVVGLRARALRGSLRTLRRMARALAATLRVTTRRLAPDRAVAEVHILKLADPQQSMELRIAVMGANEAGKSTLIGVLTQGELDNGRGSARLNMFRHLHEVKSGRTSSLSHEILGFDAQGNVVNYGCSELMTAERIGERSTRLVSFIDLAGHSKYQRTTLHGLAGYSPHFVMLVISATAGITRITEEHIDLLLALDVPFFTVVTKCELAAAAVQDVAAKLAKLLQPYNKRPLLITDENSARNCVAPQKLILDTINDNASEDKCVMPEVAVFPVSCVRGAGLGPLHACLASLACLLATPSAAHYNTLPHDDTCEFQIDEIFHAGESSGPVVGGLLARGRIREGDDLLMGPLDDGRFVSVRVVGVHRNRAAVSAARAGHTCSLRLRAQQAPRPGMVLLRQPSDSSGLHCKSNNVGSSVEDAMTLARAQQTLSKPDADATKNTFEGATDDIQDDGTTNPRGCLYFQASIHLLRHSTAIYRGFQCTVHVGNVTQTAIIQGIMSGTGELRAGESASALFRFVRSPEYLVRGRRMLFTAGLGTRAIGRITQVFPHVP
Summary
Uniprot
H9JR33
A0A2W1BNE6
A0A3S2NE96
A0A212FJ47
A0A194QAH4
A0A1B6G8K8
+ More
B4JGR0 B4MC62 A0A1I8NZZ6 A0A1B0G7J8 A0A1A9WPG1 A0A0L0BXX4 T1PCL9 A0A1B0AGP6 A0A3B0JIT9 B4K711 A0A1A9XTE8 A0A1B0BZR3 A0A0M4EPR7 A0A1A9VKN4 Q86BS2 B4PVJ7 B4I492 A0A1B6DLH1 A0A0R1E4P2 A0A1B6CL06 E0VSH9 Q960F7 A0A1L8DS02 A0A1W4V513 B4G355 B3NYI7 T1DQH7 A0A1B0CTN5 A0A182PJ28 Q297W8 A0A182J1Q8 A0A182FCZ8 B4QXM0 A0A2M4A686 A0A084VXL7 A0A067RCX3 B3LWB8 W8C7M6 A0A2M3ZHN6 W5JFF5 A0A2M4BHQ5 A0A182RQD6 A0A182NA28 A0A182MW37 A0A182JW47 A0A0A1WXY5 A0A182Q877 A0A182WK43 A0A182SIK8 B4NHG0 A0A182WRZ4 A0A182KVX0 A0A182ICX5 A0A182UNL5 A0A0K8WA52 A0A182YP08 A0A1W4X715 U5EV72 A0A336L3T0 A0A1S3D6G7 A0A195FGR5 A0A2J7PRP0 Q17B50 D6W7P8 A0A195AZX9 A0A1S4F9G8 A0A182TKS4 A0A182HD35 A0A069DVJ5 A0A023F3S0 A0A224XL62 U4UM64 T1I7D2 K7JB46 A0A232F1I0 A0A1Q3FEL0 A0A1Q3FEA9 A0A1Q3FEP4 T1IIR6 F1NAG8 A0A226P998 A0A226ND10 V5FZ98 A0A3M0KHD7 A0A1V4JJW2 B0W9N1 F4WKM3 A0A2I0USS4 A0A158NIF1 A0A2D0QKW4 E2AIG0
B4JGR0 B4MC62 A0A1I8NZZ6 A0A1B0G7J8 A0A1A9WPG1 A0A0L0BXX4 T1PCL9 A0A1B0AGP6 A0A3B0JIT9 B4K711 A0A1A9XTE8 A0A1B0BZR3 A0A0M4EPR7 A0A1A9VKN4 Q86BS2 B4PVJ7 B4I492 A0A1B6DLH1 A0A0R1E4P2 A0A1B6CL06 E0VSH9 Q960F7 A0A1L8DS02 A0A1W4V513 B4G355 B3NYI7 T1DQH7 A0A1B0CTN5 A0A182PJ28 Q297W8 A0A182J1Q8 A0A182FCZ8 B4QXM0 A0A2M4A686 A0A084VXL7 A0A067RCX3 B3LWB8 W8C7M6 A0A2M3ZHN6 W5JFF5 A0A2M4BHQ5 A0A182RQD6 A0A182NA28 A0A182MW37 A0A182JW47 A0A0A1WXY5 A0A182Q877 A0A182WK43 A0A182SIK8 B4NHG0 A0A182WRZ4 A0A182KVX0 A0A182ICX5 A0A182UNL5 A0A0K8WA52 A0A182YP08 A0A1W4X715 U5EV72 A0A336L3T0 A0A1S3D6G7 A0A195FGR5 A0A2J7PRP0 Q17B50 D6W7P8 A0A195AZX9 A0A1S4F9G8 A0A182TKS4 A0A182HD35 A0A069DVJ5 A0A023F3S0 A0A224XL62 U4UM64 T1I7D2 K7JB46 A0A232F1I0 A0A1Q3FEL0 A0A1Q3FEA9 A0A1Q3FEP4 T1IIR6 F1NAG8 A0A226P998 A0A226ND10 V5FZ98 A0A3M0KHD7 A0A1V4JJW2 B0W9N1 F4WKM3 A0A2I0USS4 A0A158NIF1 A0A2D0QKW4 E2AIG0
Pubmed
19121390
28756777
22118469
26354079
17994087
18057021
+ More
26108605 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20566863 15632085 23185243 24438588 24845553 24495485 20920257 23761445 25830018 20966253 25244985 17510324 18362917 19820115 26483478 26334808 25474469 23537049 20075255 28648823 15592404 24621616 21719571 21347285 20798317
26108605 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20566863 15632085 23185243 24438588 24845553 24495485 20920257 23761445 25830018 20966253 25244985 17510324 18362917 19820115 26483478 26334808 25474469 23537049 20075255 28648823 15592404 24621616 21719571 21347285 20798317
EMBL
BABH01028708
BABH01028709
KZ150008
PZC75154.1
RSAL01000168
RVE45222.1
+ More
AGBW02008309 OWR53777.1 KQ459463 KPJ00416.1 GECZ01011020 JAS58749.1 CH916369 EDV92664.1 CH940656 EDW58683.1 KRF78398.1 KRF78399.1 CCAG010018811 JRES01001168 KNC24878.1 KA646507 AFP61136.1 OUUW01000005 SPP80242.1 CH933806 EDW15298.1 KRG01507.1 JXJN01023274 CP012526 ALC46160.1 AE014297 AAF51934.1 AAF51936.1 AAN14330.1 ACL83471.1 CM000160 EDW97806.1 CH480821 EDW55035.1 GEDC01010810 JAS26488.1 KRK03981.1 KRK03982.1 KRK03983.1 GEDC01023149 JAS14149.1 DS235751 EEB16335.1 AY052084 AAF51935.2 AAK93508.1 GFDF01004828 JAV09256.1 CH479179 EDW24250.1 CH954181 EDV47968.1 GAMD01001150 JAB00441.1 AJWK01027789 CM000070 EAL28087.1 KRT00432.1 KRT00433.1 CM000364 EDX11806.1 GGFK01002944 MBW36265.1 ATLV01018097 KE525212 KFB42711.1 KK852543 KDR21701.1 CH902617 EDV43751.1 KPU80441.1 GAMC01003529 GAMC01003528 GAMC01003527 JAC03027.1 GGFM01007270 MBW28021.1 ADMH02001370 ETN62806.1 GGFJ01003350 MBW52491.1 AXCM01001615 GBXI01010766 JAD03526.1 AXCN02001953 CH964272 EDW84636.1 APCN01006058 GDHF01023344 GDHF01013293 GDHF01004278 GDHF01003271 GDHF01001409 JAI28970.1 JAI39021.1 JAI48036.1 JAI49043.1 JAI50905.1 GANO01001990 JAB57881.1 UFQS01001617 UFQT01001617 UFQT01001665 SSX11623.1 SSX31190.1 SSX31358.1 KQ981625 KYN39189.1 NEVH01021955 PNF19002.1 CH477325 EAT43518.1 KQ971307 EFA11327.1 KQ976698 KYM77509.1 JXUM01005146 KQ560187 KXJ83938.1 GBGD01001063 JAC87826.1 GBBI01002657 JAC16055.1 GFTR01007084 JAW09342.1 KB632384 ERL94207.1 ACPB03015441 AAZX01001765 NNAY01001262 OXU24574.1 GFDL01009061 JAV25984.1 GFDL01009144 JAV25901.1 GFDL01009043 JAV26002.1 JH430212 AADN05000005 AWGT02000117 OXB76593.1 MCFN01000094 OXB65418.1 GALX01005451 JAB63015.1 QRBI01000106 RMC12648.1 LSYS01007194 OPJ72473.1 DS231865 EDS40350.1 GL888203 EGI65254.1 KZ505642 PKU49104.1 ADTU01016768 GL439791 EFN66750.1
AGBW02008309 OWR53777.1 KQ459463 KPJ00416.1 GECZ01011020 JAS58749.1 CH916369 EDV92664.1 CH940656 EDW58683.1 KRF78398.1 KRF78399.1 CCAG010018811 JRES01001168 KNC24878.1 KA646507 AFP61136.1 OUUW01000005 SPP80242.1 CH933806 EDW15298.1 KRG01507.1 JXJN01023274 CP012526 ALC46160.1 AE014297 AAF51934.1 AAF51936.1 AAN14330.1 ACL83471.1 CM000160 EDW97806.1 CH480821 EDW55035.1 GEDC01010810 JAS26488.1 KRK03981.1 KRK03982.1 KRK03983.1 GEDC01023149 JAS14149.1 DS235751 EEB16335.1 AY052084 AAF51935.2 AAK93508.1 GFDF01004828 JAV09256.1 CH479179 EDW24250.1 CH954181 EDV47968.1 GAMD01001150 JAB00441.1 AJWK01027789 CM000070 EAL28087.1 KRT00432.1 KRT00433.1 CM000364 EDX11806.1 GGFK01002944 MBW36265.1 ATLV01018097 KE525212 KFB42711.1 KK852543 KDR21701.1 CH902617 EDV43751.1 KPU80441.1 GAMC01003529 GAMC01003528 GAMC01003527 JAC03027.1 GGFM01007270 MBW28021.1 ADMH02001370 ETN62806.1 GGFJ01003350 MBW52491.1 AXCM01001615 GBXI01010766 JAD03526.1 AXCN02001953 CH964272 EDW84636.1 APCN01006058 GDHF01023344 GDHF01013293 GDHF01004278 GDHF01003271 GDHF01001409 JAI28970.1 JAI39021.1 JAI48036.1 JAI49043.1 JAI50905.1 GANO01001990 JAB57881.1 UFQS01001617 UFQT01001617 UFQT01001665 SSX11623.1 SSX31190.1 SSX31358.1 KQ981625 KYN39189.1 NEVH01021955 PNF19002.1 CH477325 EAT43518.1 KQ971307 EFA11327.1 KQ976698 KYM77509.1 JXUM01005146 KQ560187 KXJ83938.1 GBGD01001063 JAC87826.1 GBBI01002657 JAC16055.1 GFTR01007084 JAW09342.1 KB632384 ERL94207.1 ACPB03015441 AAZX01001765 NNAY01001262 OXU24574.1 GFDL01009061 JAV25984.1 GFDL01009144 JAV25901.1 GFDL01009043 JAV26002.1 JH430212 AADN05000005 AWGT02000117 OXB76593.1 MCFN01000094 OXB65418.1 GALX01005451 JAB63015.1 QRBI01000106 RMC12648.1 LSYS01007194 OPJ72473.1 DS231865 EDS40350.1 GL888203 EGI65254.1 KZ505642 PKU49104.1 ADTU01016768 GL439791 EFN66750.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000001070
UP000008792
+ More
UP000095300 UP000092444 UP000091820 UP000037069 UP000095301 UP000092445 UP000268350 UP000009192 UP000092443 UP000092460 UP000092553 UP000078200 UP000000803 UP000002282 UP000001292 UP000009046 UP000192221 UP000008744 UP000008711 UP000092461 UP000075885 UP000001819 UP000075880 UP000069272 UP000000304 UP000030765 UP000027135 UP000007801 UP000000673 UP000075900 UP000075884 UP000075883 UP000075881 UP000075886 UP000075920 UP000075901 UP000007798 UP000076407 UP000075882 UP000075840 UP000075903 UP000076408 UP000192223 UP000079169 UP000078541 UP000235965 UP000008820 UP000007266 UP000078540 UP000075902 UP000069940 UP000249989 UP000030742 UP000015103 UP000002358 UP000215335 UP000000539 UP000198419 UP000198323 UP000269221 UP000190648 UP000002320 UP000007755 UP000005205 UP000221080 UP000000311
UP000095300 UP000092444 UP000091820 UP000037069 UP000095301 UP000092445 UP000268350 UP000009192 UP000092443 UP000092460 UP000092553 UP000078200 UP000000803 UP000002282 UP000001292 UP000009046 UP000192221 UP000008744 UP000008711 UP000092461 UP000075885 UP000001819 UP000075880 UP000069272 UP000000304 UP000030765 UP000027135 UP000007801 UP000000673 UP000075900 UP000075884 UP000075883 UP000075881 UP000075886 UP000075920 UP000075901 UP000007798 UP000076407 UP000075882 UP000075840 UP000075903 UP000076408 UP000192223 UP000079169 UP000078541 UP000235965 UP000008820 UP000007266 UP000078540 UP000075902 UP000069940 UP000249989 UP000030742 UP000015103 UP000002358 UP000215335 UP000000539 UP000198419 UP000198323 UP000269221 UP000190648 UP000002320 UP000007755 UP000005205 UP000221080 UP000000311
Pfam
PF00009 GTP_EFTU
Interpro
ProteinModelPortal
H9JR33
A0A2W1BNE6
A0A3S2NE96
A0A212FJ47
A0A194QAH4
A0A1B6G8K8
+ More
B4JGR0 B4MC62 A0A1I8NZZ6 A0A1B0G7J8 A0A1A9WPG1 A0A0L0BXX4 T1PCL9 A0A1B0AGP6 A0A3B0JIT9 B4K711 A0A1A9XTE8 A0A1B0BZR3 A0A0M4EPR7 A0A1A9VKN4 Q86BS2 B4PVJ7 B4I492 A0A1B6DLH1 A0A0R1E4P2 A0A1B6CL06 E0VSH9 Q960F7 A0A1L8DS02 A0A1W4V513 B4G355 B3NYI7 T1DQH7 A0A1B0CTN5 A0A182PJ28 Q297W8 A0A182J1Q8 A0A182FCZ8 B4QXM0 A0A2M4A686 A0A084VXL7 A0A067RCX3 B3LWB8 W8C7M6 A0A2M3ZHN6 W5JFF5 A0A2M4BHQ5 A0A182RQD6 A0A182NA28 A0A182MW37 A0A182JW47 A0A0A1WXY5 A0A182Q877 A0A182WK43 A0A182SIK8 B4NHG0 A0A182WRZ4 A0A182KVX0 A0A182ICX5 A0A182UNL5 A0A0K8WA52 A0A182YP08 A0A1W4X715 U5EV72 A0A336L3T0 A0A1S3D6G7 A0A195FGR5 A0A2J7PRP0 Q17B50 D6W7P8 A0A195AZX9 A0A1S4F9G8 A0A182TKS4 A0A182HD35 A0A069DVJ5 A0A023F3S0 A0A224XL62 U4UM64 T1I7D2 K7JB46 A0A232F1I0 A0A1Q3FEL0 A0A1Q3FEA9 A0A1Q3FEP4 T1IIR6 F1NAG8 A0A226P998 A0A226ND10 V5FZ98 A0A3M0KHD7 A0A1V4JJW2 B0W9N1 F4WKM3 A0A2I0USS4 A0A158NIF1 A0A2D0QKW4 E2AIG0
B4JGR0 B4MC62 A0A1I8NZZ6 A0A1B0G7J8 A0A1A9WPG1 A0A0L0BXX4 T1PCL9 A0A1B0AGP6 A0A3B0JIT9 B4K711 A0A1A9XTE8 A0A1B0BZR3 A0A0M4EPR7 A0A1A9VKN4 Q86BS2 B4PVJ7 B4I492 A0A1B6DLH1 A0A0R1E4P2 A0A1B6CL06 E0VSH9 Q960F7 A0A1L8DS02 A0A1W4V513 B4G355 B3NYI7 T1DQH7 A0A1B0CTN5 A0A182PJ28 Q297W8 A0A182J1Q8 A0A182FCZ8 B4QXM0 A0A2M4A686 A0A084VXL7 A0A067RCX3 B3LWB8 W8C7M6 A0A2M3ZHN6 W5JFF5 A0A2M4BHQ5 A0A182RQD6 A0A182NA28 A0A182MW37 A0A182JW47 A0A0A1WXY5 A0A182Q877 A0A182WK43 A0A182SIK8 B4NHG0 A0A182WRZ4 A0A182KVX0 A0A182ICX5 A0A182UNL5 A0A0K8WA52 A0A182YP08 A0A1W4X715 U5EV72 A0A336L3T0 A0A1S3D6G7 A0A195FGR5 A0A2J7PRP0 Q17B50 D6W7P8 A0A195AZX9 A0A1S4F9G8 A0A182TKS4 A0A182HD35 A0A069DVJ5 A0A023F3S0 A0A224XL62 U4UM64 T1I7D2 K7JB46 A0A232F1I0 A0A1Q3FEL0 A0A1Q3FEA9 A0A1Q3FEP4 T1IIR6 F1NAG8 A0A226P998 A0A226ND10 V5FZ98 A0A3M0KHD7 A0A1V4JJW2 B0W9N1 F4WKM3 A0A2I0USS4 A0A158NIF1 A0A2D0QKW4 E2AIG0
Ontologies
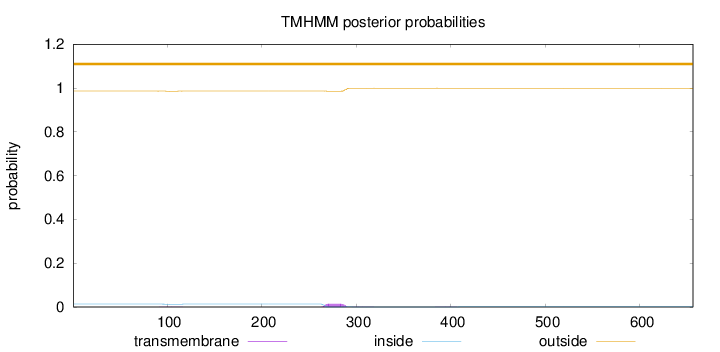
Topology
Length:
656
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.362959999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01384
outside
1 - 656
Population Genetic Test Statistics
Pi
223.77419
Theta
175.755623
Tajima's D
0.335675
CLR
1609.201617
CSRT
0.467026648667567
Interpretation
Uncertain
