Gene
KWMTBOMO06323
Annotation
pancreatic_lipase_2_[Mamestra_configurata]
Location in the cell
Extracellular Reliability : 2.322
Sequence
CDS
ATGGAGAGGTCTTGGGCGGGTTTGATTATTTTGGGCCTCGCTGCTGCTTCAGCACACAGCTATGACAATGAGGTCCTGTTAAAGCTGGAGAGCCCTCGCTACCAGCACGTCCAGGGTCCGGACGGTCTCCACCTCGAAGATCTGTGGCTAAAAGTTAAAGACCAAAAGGAGTCTGACCAATACGAGCCAGAGAGAGACAACGCATATCACTTATTCACCAGATCCAATCCTAAGACCAGCCAGCCTCTCGTGTTAGGAAGCGAAAATGTACTGAAGGCGTCTAATTACGACGAGAAAAAACGTACTATTATATTAGTACATGGATGGAGGAATGTCGCGACCTCAAACTTCAATGCTTTGCTAGTTCCGGCTTTCCTCCAGGCCGAGGATTTGAATGTCATCATCGCAGACTGGAGCATCGGGGCAGCCGGCAACTACGTAGTGGCCCTGCTGAATGTTTTTAAATCGGCCAAAGCTGTGGCTGGTTTCATCGAATGGCTGAACCGGACCAGCGGCTCCAAGGTCTCTCAGTACCACCTCGTGGGGCACAGTCTCGGCGGACACCAGGTCGGCATCATCGGGAGGCATCTGGGTGGAAAAGTTCCATACATCACTTCTCTGGACCCAGCATCACCAGGCTGGAATTTAAACCCTAACAGGTTTCAGTCTACCGACGGAATCTACACGGAGGTCATACACACTAACGCTGGGCTCAGAGGATTTCTACCCCCCCTGGGCGACATGGACTTCTATCCAAACGGTGGAGTCGAGATGCCTGGTTGCCAAGACTTAAGCTGCAGCCATCATAGATGCAGCTTCTACTTCGCCGAGTCGCTGAAGACGGGTGGATTCACCGCCCGGAGATGCCGCAACTTCGTCGAAGCCGTCAACCAGCAGTGCAACTTGCCGGAGACCGCGCAGATGGGAGGCTTACGGCCGAAAACCGGCAAAACTGGCATCTTCCATTTGCGCACAAACGCCGGATCGCCATTCTCGTTAGGATAA
Protein
MERSWAGLIILGLAAASAHSYDNEVLLKLESPRYQHVQGPDGLHLEDLWLKVKDQKESDQYEPERDNAYHLFTRSNPKTSQPLVLGSENVLKASNYDEKKRTIILVHGWRNVATSNFNALLVPAFLQAEDLNVIIADWSIGAAGNYVVALLNVFKSAKAVAGFIEWLNRTSGSKVSQYHLVGHSLGGHQVGIIGRHLGGKVPYITSLDPASPGWNLNPNRFQSTDGIYTEVIHTNAGLRGFLPPLGDMDFYPNGGVEMPGCQDLSCSHHRCSFYFAESLKTGGFTARRCRNFVEAVNQQCNLPETAQMGGLRPKTGKTGIFHLRTNAGSPFSLG
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9JQT9
A0A2A4JT49
A0A2H1VXQ6
A0A2H1VEP8
A0A2H1VLE7
A0A2W1B5Z2
+ More
A0A2W1BR39 A0A2A4K841 B2ZGH7 F5GTF9 A0A2A4K7S9 A0A2H1W3F9 A0A3S2NXP2 A0A2W1BTR8 A0A1E1W7R1 A0A1E1WHW3 A0A2H1VWT7 A0A2W1BPA5 A0A2H1V940 A0A2A4JTH4 A0A2A4IWH5 A0A2H1VU53 A0A2H1VGG1 A0A2H1VLN0 A0A0L7LGM8 B2ZGH6 A0A2W1BND5 A0A2A4JZX6 A0A2H1WKF6 B6CMF4 B1NLE7 I3QC08 A0A2W1BNC9 A0A212F9I6 A0A2H1W3D1 A0A2H1VGD1 A0A3S2NUY0 A0A3S2TIT3 A0A2A4JZ81 B6CMF6 B1NLE6 A0A1J1I2N9 I3QC06 A0A2A4JG56 A0A1W4XDL3 A0A194PKQ5 A0A2A4JEP6 I1SR90 A0A194Q4L0 A0A2A4JES0 A0A194RP46 A0A2A4K3N9 A0A194PJT7 A0A3S2M9A2 A0A2A4IX28 A0A2A4K5B8 A0A2A4JGL3 A0A194R1N2 A0A2A4IVM6 A0A068B7S7 A0A2H1V6E0 B6CMF3 A0A2A4JFT0 A0A1E1WKZ4 I3QC04 B0WAS2 A0A023PMB2 A0A2A4JFG9 I3QC05 A0A194Q2B8 A0A2H1V6Z6 A0A2H1V890 A0A182IYI0 A0A2H1WWV2 A0A212FJU4 F6K6Y6 A0A182UE51 B0WAS1 A0A182L4C6 A0A195DTF1 A0A1E1WIQ4 A0A194PVZ0 T1DJK5 A0A2A4K4B9 A0A068B851 A0A1E1VZ18 A0A1E1W907 A0A2H1V724 A0A194RKD5 A0A1E1WEQ2 A0A3S2P179 Q6L5Z4 A0A0N1PHW4 I3QC13 A0A1E1WA09 Q173Q0
A0A2W1BR39 A0A2A4K841 B2ZGH7 F5GTF9 A0A2A4K7S9 A0A2H1W3F9 A0A3S2NXP2 A0A2W1BTR8 A0A1E1W7R1 A0A1E1WHW3 A0A2H1VWT7 A0A2W1BPA5 A0A2H1V940 A0A2A4JTH4 A0A2A4IWH5 A0A2H1VU53 A0A2H1VGG1 A0A2H1VLN0 A0A0L7LGM8 B2ZGH6 A0A2W1BND5 A0A2A4JZX6 A0A2H1WKF6 B6CMF4 B1NLE7 I3QC08 A0A2W1BNC9 A0A212F9I6 A0A2H1W3D1 A0A2H1VGD1 A0A3S2NUY0 A0A3S2TIT3 A0A2A4JZ81 B6CMF6 B1NLE6 A0A1J1I2N9 I3QC06 A0A2A4JG56 A0A1W4XDL3 A0A194PKQ5 A0A2A4JEP6 I1SR90 A0A194Q4L0 A0A2A4JES0 A0A194RP46 A0A2A4K3N9 A0A194PJT7 A0A3S2M9A2 A0A2A4IX28 A0A2A4K5B8 A0A2A4JGL3 A0A194R1N2 A0A2A4IVM6 A0A068B7S7 A0A2H1V6E0 B6CMF3 A0A2A4JFT0 A0A1E1WKZ4 I3QC04 B0WAS2 A0A023PMB2 A0A2A4JFG9 I3QC05 A0A194Q2B8 A0A2H1V6Z6 A0A2H1V890 A0A182IYI0 A0A2H1WWV2 A0A212FJU4 F6K6Y6 A0A182UE51 B0WAS1 A0A182L4C6 A0A195DTF1 A0A1E1WIQ4 A0A194PVZ0 T1DJK5 A0A2A4K4B9 A0A068B851 A0A1E1VZ18 A0A1E1W907 A0A2H1V724 A0A194RKD5 A0A1E1WEQ2 A0A3S2P179 Q6L5Z4 A0A0N1PHW4 I3QC13 A0A1E1WA09 Q173Q0
Pubmed
EMBL
BABH01028718
NWSH01000654
PCG74999.1
ODYU01004911
SOQ45272.1
ODYU01002163
+ More
SOQ39305.1 ODYU01002941 SOQ41074.1 KZ150473 PZC70751.1 KZ150008 PZC75136.1 NWSH01000080 PCG79822.1 EU660854 ACD37364.1 JI484232 AEB26283.1 PCG79823.1 ODYU01006078 SOQ47611.1 RSAL01000109 RVE47195.1 PZC75133.1 GDQN01008012 JAT83042.1 GDQN01004482 JAT86572.1 SOQ45273.1 PZC75137.1 ODYU01001301 SOQ37321.1 PCG74998.1 NWSH01005705 PCG64029.1 ODYU01004467 SOQ44367.1 SOQ39304.1 ODYU01002977 SOQ41154.1 JTDY01001188 KOB74607.1 EU660853 ACD37363.1 PZC75135.1 NWSH01000332 PCG77326.1 ODYU01009267 SOQ53561.1 EU325554 ACB54944.1 EF600062 ABU98627.1 JF999957 AFI64311.1 PZC75134.1 AGBW02009585 OWR50387.1 ODYU01006077 SOQ47610.1 ODYU01002239 SOQ39452.1 RSAL01000168 RVE45226.1 RVE47196.1 PCG77325.1 EU325556 ACB54946.1 EF600061 ABU98626.1 CVRI01000038 CRK94034.1 JF999955 AFI64309.1 NWSH01001649 PCG70554.1 KQ459602 KPI93583.1 PCG70555.1 JF303743 ADZ54059.1 KQ459585 KPI98345.1 PCG70557.1 KQ460106 KPJ17796.1 NWSH01000166 PCG78875.1 KPI93582.1 RSAL01000005 RVE54443.1 PCG64026.1 PCG78872.1 PCG70553.1 KQ460870 KPJ11708.1 PCG64027.1 KJ638235 AIC80932.1 ODYU01000727 SOQ35962.1 EU325553 ACB54943.1 PCG70558.1 GDQN01003467 JAT87587.1 JF999953 AFI64307.1 DS231875 EDS41788.1 KJ439229 AHX25880.1 PCG70559.1 JF999954 AFI64308.1 KQ459589 KPI97550.1 ODYU01001007 SOQ36620.1 SOQ36622.1 ODYU01011603 SOQ57446.1 AGBW02008213 OWR54002.1 HM357823 AEA76289.1 EDS41787.1 KQ980419 KYN16118.1 GDQN01004165 JAT86889.1 KPI97551.1 GAMD01001396 JAB00195.1 PCG78876.1 KJ638234 AIC80931.1 GDQN01011122 JAT79932.1 GDQN01007598 JAT83456.1 SOQ36621.1 KPJ17795.1 GDQN01005586 JAT85468.1 RVE54445.1 AB180932 BAD22559.1 KQ460622 KPJ13522.1 JF999962 AFI64316.1 GDQN01007206 JAT83848.1 CH477418 EAT41287.1
SOQ39305.1 ODYU01002941 SOQ41074.1 KZ150473 PZC70751.1 KZ150008 PZC75136.1 NWSH01000080 PCG79822.1 EU660854 ACD37364.1 JI484232 AEB26283.1 PCG79823.1 ODYU01006078 SOQ47611.1 RSAL01000109 RVE47195.1 PZC75133.1 GDQN01008012 JAT83042.1 GDQN01004482 JAT86572.1 SOQ45273.1 PZC75137.1 ODYU01001301 SOQ37321.1 PCG74998.1 NWSH01005705 PCG64029.1 ODYU01004467 SOQ44367.1 SOQ39304.1 ODYU01002977 SOQ41154.1 JTDY01001188 KOB74607.1 EU660853 ACD37363.1 PZC75135.1 NWSH01000332 PCG77326.1 ODYU01009267 SOQ53561.1 EU325554 ACB54944.1 EF600062 ABU98627.1 JF999957 AFI64311.1 PZC75134.1 AGBW02009585 OWR50387.1 ODYU01006077 SOQ47610.1 ODYU01002239 SOQ39452.1 RSAL01000168 RVE45226.1 RVE47196.1 PCG77325.1 EU325556 ACB54946.1 EF600061 ABU98626.1 CVRI01000038 CRK94034.1 JF999955 AFI64309.1 NWSH01001649 PCG70554.1 KQ459602 KPI93583.1 PCG70555.1 JF303743 ADZ54059.1 KQ459585 KPI98345.1 PCG70557.1 KQ460106 KPJ17796.1 NWSH01000166 PCG78875.1 KPI93582.1 RSAL01000005 RVE54443.1 PCG64026.1 PCG78872.1 PCG70553.1 KQ460870 KPJ11708.1 PCG64027.1 KJ638235 AIC80932.1 ODYU01000727 SOQ35962.1 EU325553 ACB54943.1 PCG70558.1 GDQN01003467 JAT87587.1 JF999953 AFI64307.1 DS231875 EDS41788.1 KJ439229 AHX25880.1 PCG70559.1 JF999954 AFI64308.1 KQ459589 KPI97550.1 ODYU01001007 SOQ36620.1 SOQ36622.1 ODYU01011603 SOQ57446.1 AGBW02008213 OWR54002.1 HM357823 AEA76289.1 EDS41787.1 KQ980419 KYN16118.1 GDQN01004165 JAT86889.1 KPI97551.1 GAMD01001396 JAB00195.1 PCG78876.1 KJ638234 AIC80931.1 GDQN01011122 JAT79932.1 GDQN01007598 JAT83456.1 SOQ36621.1 KPJ17795.1 GDQN01005586 JAT85468.1 RVE54445.1 AB180932 BAD22559.1 KQ460622 KPJ13522.1 JF999962 AFI64316.1 GDQN01007206 JAT83848.1 CH477418 EAT41287.1
Proteomes
PRIDE
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JQT9
A0A2A4JT49
A0A2H1VXQ6
A0A2H1VEP8
A0A2H1VLE7
A0A2W1B5Z2
+ More
A0A2W1BR39 A0A2A4K841 B2ZGH7 F5GTF9 A0A2A4K7S9 A0A2H1W3F9 A0A3S2NXP2 A0A2W1BTR8 A0A1E1W7R1 A0A1E1WHW3 A0A2H1VWT7 A0A2W1BPA5 A0A2H1V940 A0A2A4JTH4 A0A2A4IWH5 A0A2H1VU53 A0A2H1VGG1 A0A2H1VLN0 A0A0L7LGM8 B2ZGH6 A0A2W1BND5 A0A2A4JZX6 A0A2H1WKF6 B6CMF4 B1NLE7 I3QC08 A0A2W1BNC9 A0A212F9I6 A0A2H1W3D1 A0A2H1VGD1 A0A3S2NUY0 A0A3S2TIT3 A0A2A4JZ81 B6CMF6 B1NLE6 A0A1J1I2N9 I3QC06 A0A2A4JG56 A0A1W4XDL3 A0A194PKQ5 A0A2A4JEP6 I1SR90 A0A194Q4L0 A0A2A4JES0 A0A194RP46 A0A2A4K3N9 A0A194PJT7 A0A3S2M9A2 A0A2A4IX28 A0A2A4K5B8 A0A2A4JGL3 A0A194R1N2 A0A2A4IVM6 A0A068B7S7 A0A2H1V6E0 B6CMF3 A0A2A4JFT0 A0A1E1WKZ4 I3QC04 B0WAS2 A0A023PMB2 A0A2A4JFG9 I3QC05 A0A194Q2B8 A0A2H1V6Z6 A0A2H1V890 A0A182IYI0 A0A2H1WWV2 A0A212FJU4 F6K6Y6 A0A182UE51 B0WAS1 A0A182L4C6 A0A195DTF1 A0A1E1WIQ4 A0A194PVZ0 T1DJK5 A0A2A4K4B9 A0A068B851 A0A1E1VZ18 A0A1E1W907 A0A2H1V724 A0A194RKD5 A0A1E1WEQ2 A0A3S2P179 Q6L5Z4 A0A0N1PHW4 I3QC13 A0A1E1WA09 Q173Q0
A0A2W1BR39 A0A2A4K841 B2ZGH7 F5GTF9 A0A2A4K7S9 A0A2H1W3F9 A0A3S2NXP2 A0A2W1BTR8 A0A1E1W7R1 A0A1E1WHW3 A0A2H1VWT7 A0A2W1BPA5 A0A2H1V940 A0A2A4JTH4 A0A2A4IWH5 A0A2H1VU53 A0A2H1VGG1 A0A2H1VLN0 A0A0L7LGM8 B2ZGH6 A0A2W1BND5 A0A2A4JZX6 A0A2H1WKF6 B6CMF4 B1NLE7 I3QC08 A0A2W1BNC9 A0A212F9I6 A0A2H1W3D1 A0A2H1VGD1 A0A3S2NUY0 A0A3S2TIT3 A0A2A4JZ81 B6CMF6 B1NLE6 A0A1J1I2N9 I3QC06 A0A2A4JG56 A0A1W4XDL3 A0A194PKQ5 A0A2A4JEP6 I1SR90 A0A194Q4L0 A0A2A4JES0 A0A194RP46 A0A2A4K3N9 A0A194PJT7 A0A3S2M9A2 A0A2A4IX28 A0A2A4K5B8 A0A2A4JGL3 A0A194R1N2 A0A2A4IVM6 A0A068B7S7 A0A2H1V6E0 B6CMF3 A0A2A4JFT0 A0A1E1WKZ4 I3QC04 B0WAS2 A0A023PMB2 A0A2A4JFG9 I3QC05 A0A194Q2B8 A0A2H1V6Z6 A0A2H1V890 A0A182IYI0 A0A2H1WWV2 A0A212FJU4 F6K6Y6 A0A182UE51 B0WAS1 A0A182L4C6 A0A195DTF1 A0A1E1WIQ4 A0A194PVZ0 T1DJK5 A0A2A4K4B9 A0A068B851 A0A1E1VZ18 A0A1E1W907 A0A2H1V724 A0A194RKD5 A0A1E1WEQ2 A0A3S2P179 Q6L5Z4 A0A0N1PHW4 I3QC13 A0A1E1WA09 Q173Q0
PDB
1GPL
E-value=2.20347e-30,
Score=329
Ontologies
GO
PANTHER
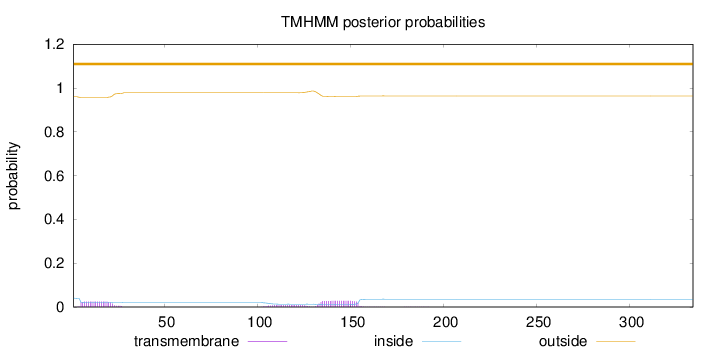
Topology
Subcellular location
Secreted
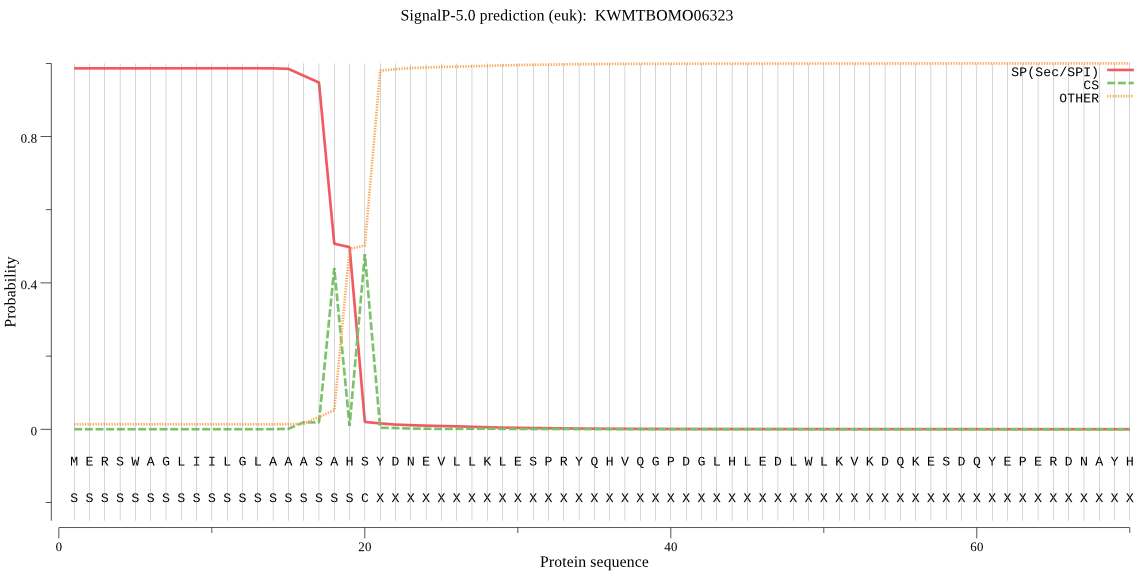
SignalP
Position: 1 - 20,
Likelihood: 0.986095
Length:
334
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.26418
Exp number, first 60 AAs:
0.45184
Total prob of N-in:
0.04031
outside
1 - 334
Population Genetic Test Statistics
Pi
277.408707
Theta
180.348612
Tajima's D
1.909846
CLR
0.057926
CSRT
0.866306684665767
Interpretation
Uncertain
