Gene
KWMTBOMO06319
Pre Gene Modal
BGIBMGA011892
Annotation
PREDICTED:_transmembrane_protein_26_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.569
Sequence
CDS
ATGTGGGTCACTGTCATCGAGCAGTTTCTGATGTTGACCCTCATTGTCGGGAGGTGGCTGCTGCCTAAGGGCGACCTGACCAGGGACCAGCTAAGTCAACTTCTCCTGGTCTACATAGGCACGGCTGCGGACATCATAGAGTTTTTCGACTCCTTCAAAGACGAGAAGGTGGCCTCCGAGAAACTCCTCGTTCTGCTCACCCTTGGCATTTGGTCGTGGTCCCTGGTGCAGTTCACCGTCGTCCTAACCGCCACAAAGTCCAGGAAGTCCCGCGGAGCAGGCAATAGGTCCGACATGAACAGCCGCGCTTGTTGCTGCACGATCGAGGTGTGCGCCATCATGATGAACATAGCTCTCCAGGACGCGCCCTTCCTGGTCTTCAGACTCCTCATAATATGCTACTACAAAATAATCAACTACATGAATATATTCTTCACCTGCAAGAACACGTTGGTCATCTTACTGCAGATGTACAGGCTATACGTGTTGCATTTGGAGATTGTTGACGACAACAATGGCGGCTCCGTGTACAGGAAGTCGGGGAGGCAGCATCACAAGAGCCGGGAGAGGAAGCCGAAGGGAGGCGGAAGAAGGTGA
Protein
MWVTVIEQFLMLTLIVGRWLLPKGDLTRDQLSQLLLVYIGTAADIIEFFDSFKDEKVASEKLLVLLTLGIWSWSLVQFTVVLTATKSRKSRGAGNRSDMNSRACCCTIEVCAIMMNIALQDAPFLVFRLLIICYYKIINYMNIFFTCKNTLVILLQMYRLYVLHLEIVDDNNGGSVYRKSGRQHHKSRERKPKGGGRR
Summary
Uniprot
H9JQT6
A0A2H1VXA3
A0A2W1BBF3
A0A0N1IFQ7
A0A3S2TIS4
A0A194Q4Y5
+ More
A0A212FD57 A0A2A4JPM0 D6WN33 A0A1W4W5S7 A0A182FNZ4 Q17BX0 A0A067QVS2 A0A182GZT8 A0A182YHH0 A0A182SXY4 A0A1A9ZA15 A0A182LUQ9 A0A182X0A0 A0A182VWL4 Q7PYR7 A0A182PFJ8 A0A232EX89 A0A182LR23 A0A182N5U4 A0A182HZY9 B0XKB0 B0X2U3 A0A182V2R9 E2BUL9 A0A310SP19 A0A151IE08 K7IMP2 A0A1B0BVV5 A0A158NSX3 A0A0L7RAT5 A0A087ZP18 A0A151XH34 A0A2A3ENE4 A0A2P8Z526 A0A195ESK0 A0A2J7QG94 A0A0M8ZXW4 A0A154PA88 B4N934 A0A0L0BKW3 W5JGE6 B4LYJ6 B4ICM3 B4K8P0 A0A1B0DH39 A0A3B0K6U9 B4PSG0 B3P670 Q9VBE7 B4G5Z2 A0A1I8MZP6 A0A1W4VKK4 B4JTL8 Q29AH4 A0A182JI60 B3M0I6 A0A336M1F4 A0A1I8PIK3 A0A336LYU1 A0A084VAG1 A0A0M4EPE7 B4QW55 A0A182H523 A0A3L8DU77 A0A026X2F4 A0A182TYW0 A0A1D2NBU7 A0A162RFH8 A0A182Q733 A0A182RC37 E9GXS5 A0A0P5BWS9 A0A1A9XRF6 A0A0P5DUS7 A0A0P5CP70 A0A0P5AYF4 A0A0P4YNM8 A0A0P6CCP7 A0A0P5YFP1 A0A0P5A3G1 A0A0P4YWG0 A0A0P6HYZ2 A0A0P5YUU9 A0A0P5PK59 A0A1J1IR81 A0A0P6EMU8 A0A226EQ56 A0A2C9JGQ2 A0A1A9UFP5 V3ZCL5 V4BNN2 A0A2C9K6N9 A0A1S3JPY4 R7UHY6
A0A212FD57 A0A2A4JPM0 D6WN33 A0A1W4W5S7 A0A182FNZ4 Q17BX0 A0A067QVS2 A0A182GZT8 A0A182YHH0 A0A182SXY4 A0A1A9ZA15 A0A182LUQ9 A0A182X0A0 A0A182VWL4 Q7PYR7 A0A182PFJ8 A0A232EX89 A0A182LR23 A0A182N5U4 A0A182HZY9 B0XKB0 B0X2U3 A0A182V2R9 E2BUL9 A0A310SP19 A0A151IE08 K7IMP2 A0A1B0BVV5 A0A158NSX3 A0A0L7RAT5 A0A087ZP18 A0A151XH34 A0A2A3ENE4 A0A2P8Z526 A0A195ESK0 A0A2J7QG94 A0A0M8ZXW4 A0A154PA88 B4N934 A0A0L0BKW3 W5JGE6 B4LYJ6 B4ICM3 B4K8P0 A0A1B0DH39 A0A3B0K6U9 B4PSG0 B3P670 Q9VBE7 B4G5Z2 A0A1I8MZP6 A0A1W4VKK4 B4JTL8 Q29AH4 A0A182JI60 B3M0I6 A0A336M1F4 A0A1I8PIK3 A0A336LYU1 A0A084VAG1 A0A0M4EPE7 B4QW55 A0A182H523 A0A3L8DU77 A0A026X2F4 A0A182TYW0 A0A1D2NBU7 A0A162RFH8 A0A182Q733 A0A182RC37 E9GXS5 A0A0P5BWS9 A0A1A9XRF6 A0A0P5DUS7 A0A0P5CP70 A0A0P5AYF4 A0A0P4YNM8 A0A0P6CCP7 A0A0P5YFP1 A0A0P5A3G1 A0A0P4YWG0 A0A0P6HYZ2 A0A0P5YUU9 A0A0P5PK59 A0A1J1IR81 A0A0P6EMU8 A0A226EQ56 A0A2C9JGQ2 A0A1A9UFP5 V3ZCL5 V4BNN2 A0A2C9K6N9 A0A1S3JPY4 R7UHY6
Pubmed
19121390
28756777
26354079
22118469
18362917
19820115
+ More
17510324 24845553 26483478 25244985 12364791 14747013 17210077 28648823 20966253 20798317 20075255 21347285 29403074 17994087 26108605 20920257 23761445 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 15632085 24438588 30249741 24508170 27289101 21292972 15562597 23254933
17510324 24845553 26483478 25244985 12364791 14747013 17210077 28648823 20966253 20798317 20075255 21347285 29403074 17994087 26108605 20920257 23761445 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 15632085 24438588 30249741 24508170 27289101 21292972 15562597 23254933
EMBL
BABH01028727
BABH01028728
BABH01028729
BABH01028730
ODYU01004842
SOQ45132.1
+ More
KZ150163 PZC72679.1 KQ460971 KPJ10121.1 RSAL01000109 RVE47173.1 KQ459463 KPJ00419.1 AGBW02009112 OWR51653.1 NWSH01000953 PCG73393.1 KQ971343 EFA04344.2 CH477315 EAT43783.1 KK853191 KDR10031.1 JXUM01002200 JXUM01002201 AXCM01006418 AAAB01008987 EAA01045.4 NNAY01001761 OXU22996.1 APCN01002023 APCN01002024 DS233765 EDS31509.1 DS232299 EDS39417.1 GL450730 EFN80606.1 KQ762329 OAD55959.1 KQ977901 KYM98911.1 JXJN01021495 ADTU01025234 ADTU01025235 ADTU01025236 ADTU01025237 ADTU01025238 ADTU01025239 ADTU01025240 ADTU01025241 ADTU01025242 ADTU01025243 KQ414618 KOC67953.1 KQ982138 KYQ59713.1 KZ288204 PBC33285.1 PYGN01000191 PSN51605.1 KQ981989 KYN31146.1 NEVH01014835 PNF27592.1 KQ435798 KOX73360.1 KQ434857 KZC08819.1 CH964232 EDW81581.1 JRES01001712 KNC20563.1 ADMH02001287 ETN63161.1 CH940650 EDW68016.1 CH480828 EDW45119.1 CH933806 EDW15459.1 AJVK01060681 OUUW01000005 SPP81356.1 CM000160 EDW98622.1 CH954182 EDV53540.1 AE014297 AAF56595.1 CH479179 EDW23924.1 CH916374 EDV91447.1 CM000070 EAL27376.2 CH902617 EDV44233.1 UFQT01000289 SSX22829.1 UFQS01000316 UFQT01000316 SSX02781.1 SSX23152.1 ATLV01003712 ATLV01003713 KE524159 KFB34955.1 CP012526 ALC47652.1 CM000364 EDX14554.1 JXUM01110793 JXUM01110794 JXUM01110795 KQ565444 KXJ70964.1 QOIP01000004 RLU23723.1 KK107020 EZA62467.1 LJIJ01000098 ODN02711.1 LRGB01000190 KZS20471.1 AXCN02001153 GL732573 EFX75766.1 GDIP01194163 JAJ29239.1 GDIP01168316 JAJ55086.1 GDIP01167426 JAJ55976.1 GDIP01192464 JAJ30938.1 GDIP01226215 JAI97186.1 GDIP01017508 JAM86207.1 GDIP01059886 JAM43829.1 GDIP01204666 JAJ18736.1 GDIP01226216 JAI97185.1 GDIQ01019926 JAN74811.1 GDIP01052785 JAM50930.1 GDIQ01137608 JAL14118.1 CVRI01000057 CRL02060.1 GDIQ01073987 JAN20750.1 LNIX01000002 OXA58951.1 KB202620 ESO88818.1 KB202408 ESO90479.1 AMQN01001274 KB300949 ELU06154.1
KZ150163 PZC72679.1 KQ460971 KPJ10121.1 RSAL01000109 RVE47173.1 KQ459463 KPJ00419.1 AGBW02009112 OWR51653.1 NWSH01000953 PCG73393.1 KQ971343 EFA04344.2 CH477315 EAT43783.1 KK853191 KDR10031.1 JXUM01002200 JXUM01002201 AXCM01006418 AAAB01008987 EAA01045.4 NNAY01001761 OXU22996.1 APCN01002023 APCN01002024 DS233765 EDS31509.1 DS232299 EDS39417.1 GL450730 EFN80606.1 KQ762329 OAD55959.1 KQ977901 KYM98911.1 JXJN01021495 ADTU01025234 ADTU01025235 ADTU01025236 ADTU01025237 ADTU01025238 ADTU01025239 ADTU01025240 ADTU01025241 ADTU01025242 ADTU01025243 KQ414618 KOC67953.1 KQ982138 KYQ59713.1 KZ288204 PBC33285.1 PYGN01000191 PSN51605.1 KQ981989 KYN31146.1 NEVH01014835 PNF27592.1 KQ435798 KOX73360.1 KQ434857 KZC08819.1 CH964232 EDW81581.1 JRES01001712 KNC20563.1 ADMH02001287 ETN63161.1 CH940650 EDW68016.1 CH480828 EDW45119.1 CH933806 EDW15459.1 AJVK01060681 OUUW01000005 SPP81356.1 CM000160 EDW98622.1 CH954182 EDV53540.1 AE014297 AAF56595.1 CH479179 EDW23924.1 CH916374 EDV91447.1 CM000070 EAL27376.2 CH902617 EDV44233.1 UFQT01000289 SSX22829.1 UFQS01000316 UFQT01000316 SSX02781.1 SSX23152.1 ATLV01003712 ATLV01003713 KE524159 KFB34955.1 CP012526 ALC47652.1 CM000364 EDX14554.1 JXUM01110793 JXUM01110794 JXUM01110795 KQ565444 KXJ70964.1 QOIP01000004 RLU23723.1 KK107020 EZA62467.1 LJIJ01000098 ODN02711.1 LRGB01000190 KZS20471.1 AXCN02001153 GL732573 EFX75766.1 GDIP01194163 JAJ29239.1 GDIP01168316 JAJ55086.1 GDIP01167426 JAJ55976.1 GDIP01192464 JAJ30938.1 GDIP01226215 JAI97186.1 GDIP01017508 JAM86207.1 GDIP01059886 JAM43829.1 GDIP01204666 JAJ18736.1 GDIP01226216 JAI97185.1 GDIQ01019926 JAN74811.1 GDIP01052785 JAM50930.1 GDIQ01137608 JAL14118.1 CVRI01000057 CRL02060.1 GDIQ01073987 JAN20750.1 LNIX01000002 OXA58951.1 KB202620 ESO88818.1 KB202408 ESO90479.1 AMQN01001274 KB300949 ELU06154.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000053268
UP000007151
UP000218220
+ More
UP000007266 UP000192223 UP000069272 UP000008820 UP000027135 UP000069940 UP000076408 UP000075901 UP000092445 UP000075883 UP000076407 UP000075920 UP000007062 UP000075885 UP000215335 UP000075882 UP000075884 UP000075840 UP000002320 UP000075903 UP000008237 UP000078542 UP000002358 UP000092460 UP000005205 UP000053825 UP000005203 UP000075809 UP000242457 UP000245037 UP000078541 UP000235965 UP000053105 UP000076502 UP000007798 UP000037069 UP000000673 UP000008792 UP000001292 UP000009192 UP000092462 UP000268350 UP000002282 UP000008711 UP000000803 UP000008744 UP000095301 UP000192221 UP000001070 UP000001819 UP000075880 UP000007801 UP000095300 UP000030765 UP000092553 UP000000304 UP000249989 UP000279307 UP000053097 UP000075902 UP000094527 UP000076858 UP000075886 UP000075900 UP000000305 UP000092443 UP000183832 UP000198287 UP000076420 UP000078200 UP000030746 UP000085678 UP000014760
UP000007266 UP000192223 UP000069272 UP000008820 UP000027135 UP000069940 UP000076408 UP000075901 UP000092445 UP000075883 UP000076407 UP000075920 UP000007062 UP000075885 UP000215335 UP000075882 UP000075884 UP000075840 UP000002320 UP000075903 UP000008237 UP000078542 UP000002358 UP000092460 UP000005205 UP000053825 UP000005203 UP000075809 UP000242457 UP000245037 UP000078541 UP000235965 UP000053105 UP000076502 UP000007798 UP000037069 UP000000673 UP000008792 UP000001292 UP000009192 UP000092462 UP000268350 UP000002282 UP000008711 UP000000803 UP000008744 UP000095301 UP000192221 UP000001070 UP000001819 UP000075880 UP000007801 UP000095300 UP000030765 UP000092553 UP000000304 UP000249989 UP000279307 UP000053097 UP000075902 UP000094527 UP000076858 UP000075886 UP000075900 UP000000305 UP000092443 UP000183832 UP000198287 UP000076420 UP000078200 UP000030746 UP000085678 UP000014760
Interpro
SUPFAM
SSF69572
SSF69572
Gene 3D
ProteinModelPortal
H9JQT6
A0A2H1VXA3
A0A2W1BBF3
A0A0N1IFQ7
A0A3S2TIS4
A0A194Q4Y5
+ More
A0A212FD57 A0A2A4JPM0 D6WN33 A0A1W4W5S7 A0A182FNZ4 Q17BX0 A0A067QVS2 A0A182GZT8 A0A182YHH0 A0A182SXY4 A0A1A9ZA15 A0A182LUQ9 A0A182X0A0 A0A182VWL4 Q7PYR7 A0A182PFJ8 A0A232EX89 A0A182LR23 A0A182N5U4 A0A182HZY9 B0XKB0 B0X2U3 A0A182V2R9 E2BUL9 A0A310SP19 A0A151IE08 K7IMP2 A0A1B0BVV5 A0A158NSX3 A0A0L7RAT5 A0A087ZP18 A0A151XH34 A0A2A3ENE4 A0A2P8Z526 A0A195ESK0 A0A2J7QG94 A0A0M8ZXW4 A0A154PA88 B4N934 A0A0L0BKW3 W5JGE6 B4LYJ6 B4ICM3 B4K8P0 A0A1B0DH39 A0A3B0K6U9 B4PSG0 B3P670 Q9VBE7 B4G5Z2 A0A1I8MZP6 A0A1W4VKK4 B4JTL8 Q29AH4 A0A182JI60 B3M0I6 A0A336M1F4 A0A1I8PIK3 A0A336LYU1 A0A084VAG1 A0A0M4EPE7 B4QW55 A0A182H523 A0A3L8DU77 A0A026X2F4 A0A182TYW0 A0A1D2NBU7 A0A162RFH8 A0A182Q733 A0A182RC37 E9GXS5 A0A0P5BWS9 A0A1A9XRF6 A0A0P5DUS7 A0A0P5CP70 A0A0P5AYF4 A0A0P4YNM8 A0A0P6CCP7 A0A0P5YFP1 A0A0P5A3G1 A0A0P4YWG0 A0A0P6HYZ2 A0A0P5YUU9 A0A0P5PK59 A0A1J1IR81 A0A0P6EMU8 A0A226EQ56 A0A2C9JGQ2 A0A1A9UFP5 V3ZCL5 V4BNN2 A0A2C9K6N9 A0A1S3JPY4 R7UHY6
A0A212FD57 A0A2A4JPM0 D6WN33 A0A1W4W5S7 A0A182FNZ4 Q17BX0 A0A067QVS2 A0A182GZT8 A0A182YHH0 A0A182SXY4 A0A1A9ZA15 A0A182LUQ9 A0A182X0A0 A0A182VWL4 Q7PYR7 A0A182PFJ8 A0A232EX89 A0A182LR23 A0A182N5U4 A0A182HZY9 B0XKB0 B0X2U3 A0A182V2R9 E2BUL9 A0A310SP19 A0A151IE08 K7IMP2 A0A1B0BVV5 A0A158NSX3 A0A0L7RAT5 A0A087ZP18 A0A151XH34 A0A2A3ENE4 A0A2P8Z526 A0A195ESK0 A0A2J7QG94 A0A0M8ZXW4 A0A154PA88 B4N934 A0A0L0BKW3 W5JGE6 B4LYJ6 B4ICM3 B4K8P0 A0A1B0DH39 A0A3B0K6U9 B4PSG0 B3P670 Q9VBE7 B4G5Z2 A0A1I8MZP6 A0A1W4VKK4 B4JTL8 Q29AH4 A0A182JI60 B3M0I6 A0A336M1F4 A0A1I8PIK3 A0A336LYU1 A0A084VAG1 A0A0M4EPE7 B4QW55 A0A182H523 A0A3L8DU77 A0A026X2F4 A0A182TYW0 A0A1D2NBU7 A0A162RFH8 A0A182Q733 A0A182RC37 E9GXS5 A0A0P5BWS9 A0A1A9XRF6 A0A0P5DUS7 A0A0P5CP70 A0A0P5AYF4 A0A0P4YNM8 A0A0P6CCP7 A0A0P5YFP1 A0A0P5A3G1 A0A0P4YWG0 A0A0P6HYZ2 A0A0P5YUU9 A0A0P5PK59 A0A1J1IR81 A0A0P6EMU8 A0A226EQ56 A0A2C9JGQ2 A0A1A9UFP5 V3ZCL5 V4BNN2 A0A2C9K6N9 A0A1S3JPY4 R7UHY6
Ontologies
PANTHER
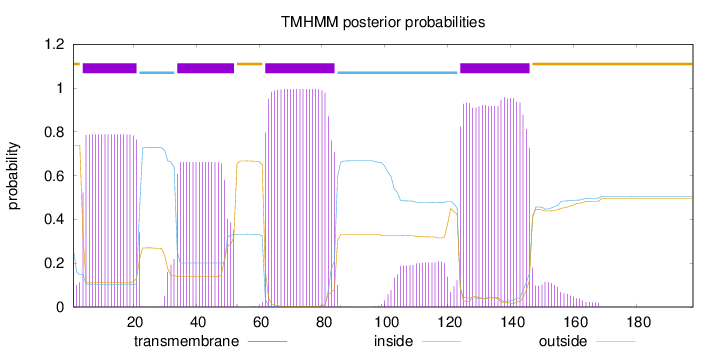
Topology
Length:
198
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
75.06454
Exp number, first 60 AAs:
26.67953
Total prob of N-in:
0.26143
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 21
inside
22 - 33
TMhelix
34 - 52
outside
53 - 61
TMhelix
62 - 84
inside
85 - 123
TMhelix
124 - 146
outside
147 - 198
Population Genetic Test Statistics
Pi
218.067846
Theta
154.145394
Tajima's D
1.33198
CLR
0
CSRT
0.744312784360782
Interpretation
Uncertain
