Pre Gene Modal
BGIBMGA011891
Annotation
PREDICTED:_adenylyltransferase_and_sulfurtransferase_MOCS3_isoform_X1_[Papilio_xuthus]
Full name
Adenylyltransferase and sulfurtransferase MOCS3 homolog
+ More
Adenylyltransferase and sulfurtransferase MOCS3
Adenylyltransferase and sulfurtransferase UBA4
Adenylyltransferase and sulfurtransferase MOCS3
Adenylyltransferase and sulfurtransferase UBA4
Alternative Name
UBA4 homolog
Ubiquitin-like protein activator 4 homolog
Molybdenum cofactor synthesis protein 3
Ubiquitin activating enzyme 4
Ubiquitin-like protein activator 4
Ubiquitin-like protein activator 4 homolog
Molybdenum cofactor synthesis protein 3
Ubiquitin activating enzyme 4
Ubiquitin-like protein activator 4
Location in the cell
Cytoplasmic Reliability : 1.928 Nuclear Reliability : 1.424
Sequence
CDS
ATGGACAGAGTCAGTGTTTTAGAGACGGAAATATCAGAACTTCGCAGAATTTTAAAGGAAAAGGAAAATGAACTTCAGGAATTAAAAAGTTTGATTAGTGATCAACAATCAAGTAAAGAATTAAATAATCACAGTCAGCCGATAAGTGATGCTTCAAAATCATCAGTAGATGCCTGTCTTCCAAAATGGGCGATAGAAAGGTACAGTCGTCAGATCCTTCTCTCGGACATCGGCGTCGAAGGACAGGTTAAGATCTGTTCTGCCAAAGTACTGATTGTGGGAGCCGGCGGCCTGGGATGTCCGGCTGCCATGTACCTTGCCGGGGCGGGCATTGGCGAAATAGGCATCGTGGATTATGACGCGGTGGATCTGACCAACGTGCACAGACAACTGCTGCACCATGAAAGCAACGAGAACACCAGCAAGGCGTTCTCGGCCGCTGAGTCTCTGAGATGTATAAACTCGAAAATTAAAATCACACCATACAACGTTCAGTTGGACTCGAAGAACGCTCAGCAAATCGCTTCAGCTTATGACTTGGTGCTCGACTGCAGCGACAACGTCCCGACCAGGTATCTGTTGAACGACCTGTGCGTTATGCTAAAGATCCCTCTGATATCGGGAAGCGCTTTGAAAATGGAGGGGCAGCTCACGATATACGGGTACAGGAGACAGGACCATCACCAGGAGACGTACATAGGGCCTTGCTACCGCTGCGTGTTCCCCACGCCCCCCCCTCCCGCTGCGGTGGGCAGCTGCTCCGCCCACGGGGTGGCCGGGCCCGTCCCGGGGACCGTGGGAACCCTGCAGGCCCTGGAGGCGATAAAGTACGTGATAGGCAGGACCGAGAGGCACCTGCTGGTCGGCCGCATGCTGCTGTTCGACGGAGAGGACGCGACCTTTAGGGCAGTAAAGTTGCGTCCGAGGAGACCCGCGTGCGTGGCGTGTGGGGACCGCGGAGGGGGCCGCCTGATCGATTATGAGCAGTTCTGCAATGCGCCGGCCAAAGAGAAGGACTTGGATCTGGAGGTGCTGCCGCCGGAAAATCGGGTCAGCGCCAACGAACTGAACGTAATGATGAATAACGAGGAGCGAGGAGGACGTCATCTCGTGGTGGACGTTAGGAACGAGCAGGAGTACGCGATGTGCCACATTCAGGGCTCGGTCAACTACCCCGTGGAGAACTTGCACCGGAACTTGGAGGAGTTGATCGAGAAGACCAAGGAGTTTGATGGAGGTGAGCTTTTTTTTATTGCTTAA
Protein
MDRVSVLETEISELRRILKEKENELQELKSLISDQQSSKELNNHSQPISDASKSSVDACLPKWAIERYSRQILLSDIGVEGQVKICSAKVLIVGAGGLGCPAAMYLAGAGIGEIGIVDYDAVDLTNVHRQLLHHESNENTSKAFSAAESLRCINSKIKITPYNVQLDSKNAQQIASAYDLVLDCSDNVPTRYLLNDLCVMLKIPLISGSALKMEGQLTIYGYRRQDHHQETYIGPCYRCVFPTPPPPAAVGSCSAHGVAGPVPGTVGTLQALEAIKYVIGRTERHLLVGRMLLFDGEDATFRAVKLRPRRPACVACGDRGGGRLIDYEQFCNAPAKEKDLDLEVLPPENRVSANELNVMMNNEERGGRHLVVDVRNEQEYAMCHIQGSVNYPVENLHRNLEELIEKTKEFDGGELFFIA
Summary
Description
Plays a central role in 2-thiolation of mcm(5)S(2)U at tRNA wobble positions of cytosolic tRNA(Lys), tRNA(Glu) and tRNA(Gln). Acts by mediating the C-terminal thiocarboxylation of the sulfur carrier URM1. Its N-terminus first activates URM1 as acyl-adenylate (-COAMP), then the persulfide sulfur on the catalytic cysteine is transferred to URM1 to form thiocarboxylation (-COSH) of its C-terminus. The reaction probably involves hydrogen sulfide that is generated from the persulfide intermediate and that acts as nucleophile towards URM1. Subsequently, a transient disulfide bond is formed. Does not use thiosulfate as sulfur donor; NFS1 probably acting as a sulfur donor for thiocarboxylation reactions.
Plays a central role in 2-thiolation of mcm(5)S(2)U at tRNA wobble positions of cytosolic tRNA(Lys), tRNA(Glu) and tRNA(Gln). Also essential during biosynthesis of the molybdenum cofactor. Acts by mediating the C-terminal thiocarboxylation of sulfur carriers URM1 and MOCS2A. Its N-terminus first activates URM1 and MOCS2A as acyl-adenylates (-COAMP), then the persulfide sulfur on the catalytic cysteine is transferred to URM1 and MOCS2A to form thiocarboxylation (-COSH) of their C-terminus. The reaction probably involves hydrogen sulfide that is generated from the persulfide intermediate and that acts as nucleophile towards URM1 and MOCS2A. Subsequently, a transient disulfide bond is formed. Does not use thiosulfate as sulfur donor; NFS1 probably acting as a sulfur donor for thiocarboxylation reactions.
Plays a central role in 2-thiolation of mcm(5)S(2)U at tRNA wobble positions of cytosolic tRNA(Lys), tRNA(Glu) and tRNA(Gln). Acts by mediating the C-terminal thiocarboxylation of sulfur carrier URM1. Its N-terminus first activates URM1 as acyl-adenylate (-COAMP), then the persulfide sulfur on the catalytic cysteine is transferred to URM1 to form thiocarboxylation (-COSH) of its C-terminus. The reaction probably involves hydrogen sulfide that is generated from the persulfide intermediate and that acts as nucleophile towards URM1. Subsequently, a transient disulfide bond is formed. Does not use thiosulfate as sulfur donor; NFS1 probably acting as a sulfur donor for thiocarboxylation reactions. Prior mcm(5) tRNA modification by the elongator complex is required for 2-thiolation. May also be involved in protein urmylation.
Plays a central role in 2-thiolation of mcm(5)S(2)U at tRNA wobble positions of cytosolic tRNA(Lys), tRNA(Glu) and tRNA(Gln). Also essential during biosynthesis of the molybdenum cofactor. Acts by mediating the C-terminal thiocarboxylation of sulfur carriers URM1 and MOCS2A. Its N-terminus first activates URM1 and MOCS2A as acyl-adenylates (-COAMP), then the persulfide sulfur on the catalytic cysteine is transferred to URM1 and MOCS2A to form thiocarboxylation (-COSH) of their C-terminus. The reaction probably involves hydrogen sulfide that is generated from the persulfide intermediate and that acts as nucleophile towards URM1 and MOCS2A. Subsequently, a transient disulfide bond is formed. Does not use thiosulfate as sulfur donor; NFS1 probably acting as a sulfur donor for thiocarboxylation reactions.
Plays a central role in 2-thiolation of mcm(5)S(2)U at tRNA wobble positions of cytosolic tRNA(Lys), tRNA(Glu) and tRNA(Gln). Acts by mediating the C-terminal thiocarboxylation of sulfur carrier URM1. Its N-terminus first activates URM1 as acyl-adenylate (-COAMP), then the persulfide sulfur on the catalytic cysteine is transferred to URM1 to form thiocarboxylation (-COSH) of its C-terminus. The reaction probably involves hydrogen sulfide that is generated from the persulfide intermediate and that acts as nucleophile towards URM1. Subsequently, a transient disulfide bond is formed. Does not use thiosulfate as sulfur donor; NFS1 probably acting as a sulfur donor for thiocarboxylation reactions. Prior mcm(5) tRNA modification by the elongator complex is required for 2-thiolation. May also be involved in protein urmylation.
Catalytic Activity
[molybdopterin-synthase sulfur-carrier protein]-C-terminal Gly-Gly + ATP + H(+) = [molybdopterin-synthase sulfur-carrier protein]-C-terminal Gly-Gly-AMP + diphosphate
[molybdopterin-synthase sulfur-carrier protein]-C-terminal Gly-Gly-AMP + AH2 + S-sulfanyl-L-cysteinyl-[cysteine desulfurase] = [molybdopterin-synthase sulfur-carrier protein]-C-terminal Gly-NH-CH2-C(O)SH + A + AMP + H(+) + L-cysteinyl-[cysteine desulfurase]
[molybdopterin-synthase sulfur-carrier protein]-C-terminal Gly-Gly-AMP + AH2 + S-sulfanyl-L-cysteinyl-[cysteine desulfurase] = [molybdopterin-synthase sulfur-carrier protein]-C-terminal Gly-NH-CH2-C(O)SH + A + AMP + H(+) + L-cysteinyl-[cysteine desulfurase]
Cofactor
Zn(2+)
Similarity
In the N-terminal section; belongs to the HesA/MoeB/ThiF family. UBA4 subfamily.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Metal-binding
Molybdenum cofactor biosynthesis
Multifunctional enzyme
Nucleotide-binding
Reference proteome
Transferase
tRNA processing
Zinc
Feature
chain Adenylyltransferase and sulfurtransferase MOCS3
Uniprot
H9JQT5
A0A3S2NXN3
A0A2A4K967
A0A2H1W8W7
A0A182FPX2
U5EW75
+ More
B0W377 A0A2W1BD56 A0A182Y3U8 A0A182RSX2 A0A182M1E2 A0A182GLG4 A0A182H1U8 A0A1Y9IVZ6 A0A023EUI2 A0A1I8JTE7 Q7PY41 A0A182NSV6 A0A182PSH0 A0A0L7RC21 A0A182INL1 A0A1I8JVV7 A0A182JW94 A0A182U995 A0A1Q3F9S4 Q17CA7 A0A2A3E7A6 A0A154PI34 A0A2K7P699 A0A182V2V2 A0A084VZE7 A0A087ZMT7 K7J5G0 A0A158NJ76 A0A195BKU7 A0A232FKV3 A0A2M4AHX4 A0A210PXI6 A0A336M9W7 A0A2M4BQS2 A0A1B6LZH7 A0A2M4BQH9 A0A0N8A194 A0A0P5BSM7 A0A0N8BGR9 W5JI26 A0A0P5XJ57 E2AJ64 A0A0N8B6W2 A0A0P5NXT0 A0A0P6FD10 A0A1B6GAM7 A0A0P5SGE4 A0A0P4WQY5 A0A0N7ZTT7 A0A3B5BGN4 A0A0P5QWH9 A0A026WWM2 A0A164L0V4 A0A0P6ACZ8 A0A0P5I8L0 A0A0P5IUT9 A0A0P6IFC5 A0A1S3J2P2 A0A0P5W462 A0A182QNA0 A0A3B3C7A8 F4WKP0 A0A151JV91 A0A151WE67 A0A0B6ZFS4 A0A1B6IGC9 A0A151J922 A0A0K8S6F3 A0A0P4Z5M7 A0A2C9LM44 A0A151I7G9 V3ZM40 B4LRB9 A0A0A9YR34 A0A0P4VQ85 T1HJ70 A0A146L9V2 A0A1B0AKS4 A0A3Q1EXR4 A0A3B0JXX0 G3NV70 A0A1A9XUH2 E5SIM0 A0A310SGL4 A0A0V0VQR2 A0A0V1PBK0 A0A3P8UHD4 A0A0V0TYT8 A0A1Y3E887 G3NV64 A0A3P8UIC5 A0A177WB71 F4NU54
B0W377 A0A2W1BD56 A0A182Y3U8 A0A182RSX2 A0A182M1E2 A0A182GLG4 A0A182H1U8 A0A1Y9IVZ6 A0A023EUI2 A0A1I8JTE7 Q7PY41 A0A182NSV6 A0A182PSH0 A0A0L7RC21 A0A182INL1 A0A1I8JVV7 A0A182JW94 A0A182U995 A0A1Q3F9S4 Q17CA7 A0A2A3E7A6 A0A154PI34 A0A2K7P699 A0A182V2V2 A0A084VZE7 A0A087ZMT7 K7J5G0 A0A158NJ76 A0A195BKU7 A0A232FKV3 A0A2M4AHX4 A0A210PXI6 A0A336M9W7 A0A2M4BQS2 A0A1B6LZH7 A0A2M4BQH9 A0A0N8A194 A0A0P5BSM7 A0A0N8BGR9 W5JI26 A0A0P5XJ57 E2AJ64 A0A0N8B6W2 A0A0P5NXT0 A0A0P6FD10 A0A1B6GAM7 A0A0P5SGE4 A0A0P4WQY5 A0A0N7ZTT7 A0A3B5BGN4 A0A0P5QWH9 A0A026WWM2 A0A164L0V4 A0A0P6ACZ8 A0A0P5I8L0 A0A0P5IUT9 A0A0P6IFC5 A0A1S3J2P2 A0A0P5W462 A0A182QNA0 A0A3B3C7A8 F4WKP0 A0A151JV91 A0A151WE67 A0A0B6ZFS4 A0A1B6IGC9 A0A151J922 A0A0K8S6F3 A0A0P4Z5M7 A0A2C9LM44 A0A151I7G9 V3ZM40 B4LRB9 A0A0A9YR34 A0A0P4VQ85 T1HJ70 A0A146L9V2 A0A1B0AKS4 A0A3Q1EXR4 A0A3B0JXX0 G3NV70 A0A1A9XUH2 E5SIM0 A0A310SGL4 A0A0V0VQR2 A0A0V1PBK0 A0A3P8UHD4 A0A0V0TYT8 A0A1Y3E887 G3NV64 A0A3P8UIC5 A0A177WB71 F4NU54
Pubmed
EMBL
BABH01028732
BABH01028733
BABH01028734
RSAL01000109
RVE47171.1
NWSH01000042
+ More
PCG80333.1 ODYU01007055 SOQ49487.1 GANO01001549 JAB58322.1 DS231831 KZ150447 PZC70840.1 AXCM01000028 JXUM01071933 KQ562693 KXJ75321.1 JXUM01023254 JXUM01023255 KQ560656 KXJ81407.1 GAPW01001479 JAC12119.1 APCN01000910 AAAB01008987 KQ414617 KOC68384.1 GFDL01010749 JAV24296.1 CH477310 KZ288363 PBC26921.1 KQ434899 KZC10860.1 ATLV01018748 KE525248 KFB43341.1 ADTU01017554 KQ976453 KYM85394.1 NNAY01000065 OXU31364.1 GGFK01006887 MBW40208.1 NEDP02005416 OWF41195.1 UFQT01000609 SSX25689.1 GGFJ01006130 MBW55271.1 GEBQ01010932 JAT29045.1 GGFJ01006131 MBW55272.1 GDIP01192385 JAJ31017.1 GDIP01195434 JAJ27968.1 GDIQ01179307 GDIQ01177866 JAK72418.1 ADMH02001345 ETN62938.1 GDIP01083602 JAM20113.1 GL439967 EFN66504.1 GDIQ01206963 JAK44762.1 GDIQ01143271 JAL08455.1 GDIQ01065566 JAN29171.1 GECZ01010271 JAS59498.1 GDIP01140046 JAL63668.1 GDIP01254272 JAI69129.1 GDIP01213241 JAJ10161.1 GDIQ01115388 JAL36338.1 KK107078 QOIP01000011 EZA60457.1 RLU16629.1 LRGB01003216 KZS03699.1 GDIP01031005 JAM72710.1 GDIQ01217302 JAK34423.1 GDIQ01216308 JAK35417.1 GDIQ01006889 JAN87848.1 GDIP01091299 JAM12416.1 AXCN02001149 GL888206 EGI65132.1 KQ981703 KYN37443.1 KQ983247 KYQ46164.1 HACG01019831 CEK66696.1 GECU01021730 JAS85976.1 KQ979480 KYN21255.1 GBRD01017083 JAG48744.1 GDIP01221947 JAJ01455.1 KQ978411 KYM94096.1 KB203660 ESO83505.1 CH940649 GBHO01008965 JAG34639.1 GDKW01002604 JAI53991.1 ACPB03021235 GDHC01015219 JAQ03410.1 JXJN01027914 OUUW01000010 SPP85913.1 JYDH01000114 KRY31641.1 KQ764145 OAD54590.1 JYDN01000016 KRX65501.1 JYDM01000024 KRZ93612.1 JYDJ01000102 KRX44186.1 LVZM01021474 OUC41342.1 DS022300 OAJ37357.1 GL882879 EGF84382.1
PCG80333.1 ODYU01007055 SOQ49487.1 GANO01001549 JAB58322.1 DS231831 KZ150447 PZC70840.1 AXCM01000028 JXUM01071933 KQ562693 KXJ75321.1 JXUM01023254 JXUM01023255 KQ560656 KXJ81407.1 GAPW01001479 JAC12119.1 APCN01000910 AAAB01008987 KQ414617 KOC68384.1 GFDL01010749 JAV24296.1 CH477310 KZ288363 PBC26921.1 KQ434899 KZC10860.1 ATLV01018748 KE525248 KFB43341.1 ADTU01017554 KQ976453 KYM85394.1 NNAY01000065 OXU31364.1 GGFK01006887 MBW40208.1 NEDP02005416 OWF41195.1 UFQT01000609 SSX25689.1 GGFJ01006130 MBW55271.1 GEBQ01010932 JAT29045.1 GGFJ01006131 MBW55272.1 GDIP01192385 JAJ31017.1 GDIP01195434 JAJ27968.1 GDIQ01179307 GDIQ01177866 JAK72418.1 ADMH02001345 ETN62938.1 GDIP01083602 JAM20113.1 GL439967 EFN66504.1 GDIQ01206963 JAK44762.1 GDIQ01143271 JAL08455.1 GDIQ01065566 JAN29171.1 GECZ01010271 JAS59498.1 GDIP01140046 JAL63668.1 GDIP01254272 JAI69129.1 GDIP01213241 JAJ10161.1 GDIQ01115388 JAL36338.1 KK107078 QOIP01000011 EZA60457.1 RLU16629.1 LRGB01003216 KZS03699.1 GDIP01031005 JAM72710.1 GDIQ01217302 JAK34423.1 GDIQ01216308 JAK35417.1 GDIQ01006889 JAN87848.1 GDIP01091299 JAM12416.1 AXCN02001149 GL888206 EGI65132.1 KQ981703 KYN37443.1 KQ983247 KYQ46164.1 HACG01019831 CEK66696.1 GECU01021730 JAS85976.1 KQ979480 KYN21255.1 GBRD01017083 JAG48744.1 GDIP01221947 JAJ01455.1 KQ978411 KYM94096.1 KB203660 ESO83505.1 CH940649 GBHO01008965 JAG34639.1 GDKW01002604 JAI53991.1 ACPB03021235 GDHC01015219 JAQ03410.1 JXJN01027914 OUUW01000010 SPP85913.1 JYDH01000114 KRY31641.1 KQ764145 OAD54590.1 JYDN01000016 KRX65501.1 JYDM01000024 KRZ93612.1 JYDJ01000102 KRX44186.1 LVZM01021474 OUC41342.1 DS022300 OAJ37357.1 GL882879 EGF84382.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000069272
UP000002320
UP000076408
+ More
UP000075900 UP000075883 UP000069940 UP000249989 UP000075920 UP000075840 UP000007062 UP000075884 UP000075885 UP000053825 UP000075880 UP000076407 UP000075881 UP000075902 UP000008820 UP000242457 UP000076502 UP000075903 UP000030765 UP000005203 UP000002358 UP000005205 UP000078540 UP000215335 UP000242188 UP000000673 UP000000311 UP000261400 UP000053097 UP000279307 UP000076858 UP000085678 UP000075886 UP000261560 UP000007755 UP000078541 UP000075809 UP000078492 UP000076420 UP000078542 UP000030746 UP000008792 UP000015103 UP000092460 UP000257200 UP000268350 UP000007635 UP000092443 UP000054776 UP000054681 UP000054924 UP000265120 UP000055048 UP000243006 UP000077115 UP000007241
UP000075900 UP000075883 UP000069940 UP000249989 UP000075920 UP000075840 UP000007062 UP000075884 UP000075885 UP000053825 UP000075880 UP000076407 UP000075881 UP000075902 UP000008820 UP000242457 UP000076502 UP000075903 UP000030765 UP000005203 UP000002358 UP000005205 UP000078540 UP000215335 UP000242188 UP000000673 UP000000311 UP000261400 UP000053097 UP000279307 UP000076858 UP000085678 UP000075886 UP000261560 UP000007755 UP000078541 UP000075809 UP000078492 UP000076420 UP000078542 UP000030746 UP000008792 UP000015103 UP000092460 UP000257200 UP000268350 UP000007635 UP000092443 UP000054776 UP000054681 UP000054924 UP000265120 UP000055048 UP000243006 UP000077115 UP000007241
Interpro
Gene 3D
ProteinModelPortal
H9JQT5
A0A3S2NXN3
A0A2A4K967
A0A2H1W8W7
A0A182FPX2
U5EW75
+ More
B0W377 A0A2W1BD56 A0A182Y3U8 A0A182RSX2 A0A182M1E2 A0A182GLG4 A0A182H1U8 A0A1Y9IVZ6 A0A023EUI2 A0A1I8JTE7 Q7PY41 A0A182NSV6 A0A182PSH0 A0A0L7RC21 A0A182INL1 A0A1I8JVV7 A0A182JW94 A0A182U995 A0A1Q3F9S4 Q17CA7 A0A2A3E7A6 A0A154PI34 A0A2K7P699 A0A182V2V2 A0A084VZE7 A0A087ZMT7 K7J5G0 A0A158NJ76 A0A195BKU7 A0A232FKV3 A0A2M4AHX4 A0A210PXI6 A0A336M9W7 A0A2M4BQS2 A0A1B6LZH7 A0A2M4BQH9 A0A0N8A194 A0A0P5BSM7 A0A0N8BGR9 W5JI26 A0A0P5XJ57 E2AJ64 A0A0N8B6W2 A0A0P5NXT0 A0A0P6FD10 A0A1B6GAM7 A0A0P5SGE4 A0A0P4WQY5 A0A0N7ZTT7 A0A3B5BGN4 A0A0P5QWH9 A0A026WWM2 A0A164L0V4 A0A0P6ACZ8 A0A0P5I8L0 A0A0P5IUT9 A0A0P6IFC5 A0A1S3J2P2 A0A0P5W462 A0A182QNA0 A0A3B3C7A8 F4WKP0 A0A151JV91 A0A151WE67 A0A0B6ZFS4 A0A1B6IGC9 A0A151J922 A0A0K8S6F3 A0A0P4Z5M7 A0A2C9LM44 A0A151I7G9 V3ZM40 B4LRB9 A0A0A9YR34 A0A0P4VQ85 T1HJ70 A0A146L9V2 A0A1B0AKS4 A0A3Q1EXR4 A0A3B0JXX0 G3NV70 A0A1A9XUH2 E5SIM0 A0A310SGL4 A0A0V0VQR2 A0A0V1PBK0 A0A3P8UHD4 A0A0V0TYT8 A0A1Y3E887 G3NV64 A0A3P8UIC5 A0A177WB71 F4NU54
B0W377 A0A2W1BD56 A0A182Y3U8 A0A182RSX2 A0A182M1E2 A0A182GLG4 A0A182H1U8 A0A1Y9IVZ6 A0A023EUI2 A0A1I8JTE7 Q7PY41 A0A182NSV6 A0A182PSH0 A0A0L7RC21 A0A182INL1 A0A1I8JVV7 A0A182JW94 A0A182U995 A0A1Q3F9S4 Q17CA7 A0A2A3E7A6 A0A154PI34 A0A2K7P699 A0A182V2V2 A0A084VZE7 A0A087ZMT7 K7J5G0 A0A158NJ76 A0A195BKU7 A0A232FKV3 A0A2M4AHX4 A0A210PXI6 A0A336M9W7 A0A2M4BQS2 A0A1B6LZH7 A0A2M4BQH9 A0A0N8A194 A0A0P5BSM7 A0A0N8BGR9 W5JI26 A0A0P5XJ57 E2AJ64 A0A0N8B6W2 A0A0P5NXT0 A0A0P6FD10 A0A1B6GAM7 A0A0P5SGE4 A0A0P4WQY5 A0A0N7ZTT7 A0A3B5BGN4 A0A0P5QWH9 A0A026WWM2 A0A164L0V4 A0A0P6ACZ8 A0A0P5I8L0 A0A0P5IUT9 A0A0P6IFC5 A0A1S3J2P2 A0A0P5W462 A0A182QNA0 A0A3B3C7A8 F4WKP0 A0A151JV91 A0A151WE67 A0A0B6ZFS4 A0A1B6IGC9 A0A151J922 A0A0K8S6F3 A0A0P4Z5M7 A0A2C9LM44 A0A151I7G9 V3ZM40 B4LRB9 A0A0A9YR34 A0A0P4VQ85 T1HJ70 A0A146L9V2 A0A1B0AKS4 A0A3Q1EXR4 A0A3B0JXX0 G3NV70 A0A1A9XUH2 E5SIM0 A0A310SGL4 A0A0V0VQR2 A0A0V1PBK0 A0A3P8UHD4 A0A0V0TYT8 A0A1Y3E887 G3NV64 A0A3P8UIC5 A0A177WB71 F4NU54
PDB
1JWB
E-value=4.57211e-43,
Score=440
Ontologies
GO
GO:0005524
GO:0006777
GO:0018192
GO:0008641
GO:0046872
GO:0005829
GO:0002143
GO:0004792
GO:0016779
GO:0034227
GO:0016783
GO:0002098
GO:0005737
GO:0016740
GO:0061604
GO:0061605
GO:0042292
GO:0032447
GO:0016021
GO:0032324
GO:0019008
GO:0051015
GO:0045296
GO:0016020
GO:0016627
GO:0030145
GO:0015923
GO:0005634
GO:0006260
Topology
Subcellular location
Cytoplasm
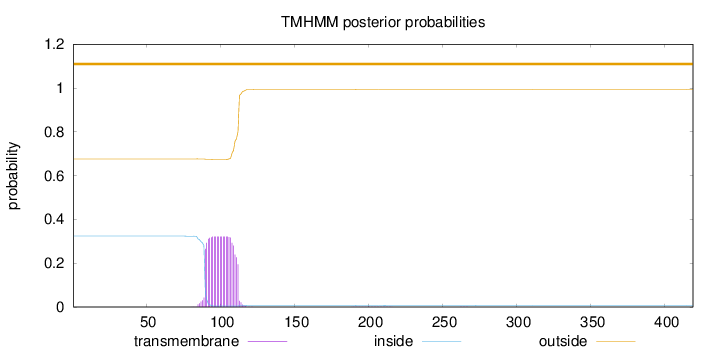
Length:
419
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.13122
Exp number, first 60 AAs:
0
Total prob of N-in:
0.32372
outside
1 - 419
Population Genetic Test Statistics
Pi
251.413426
Theta
173.40865
Tajima's D
1.188086
CLR
0.112817
CSRT
0.708964551772411
Interpretation
Uncertain
