Pre Gene Modal
BGIBMGA011887
Annotation
PREDICTED:_dihydroorotate_dehydrogenase_(quinone)?_mitochondrial_isoform_X1_[Amyelois_transitella]
Full name
Dihydroorotate dehydrogenase (quinone), mitochondrial
Location in the cell
Cytoplasmic Reliability : 1.426 Mitochondrial Reliability : 1.925
Sequence
CDS
ATGTCGCAGAAGAAAGACCCTTTGAAAAAAATTAGATCCCTTTGTTATGTGACAATCGGAGGAGCTATCGCCTACCAGTATATTTATTACAAGAAGGACTTTAACAGTTATTACGAGAACGTCCTCCAACCTCTCAGTCAGTATCTGAGTCCTGAAGTCGCTCATCGAATCGGAGTGGCAGCGATCAAACACGGACTATTTCCACCTGATCAAAATGAAGACCCAAAGATTTTAAAAACAAGATTGCTTGACTACGACCTAAGCAATCCACTTGGCATAGCCGCCGGGTTCGACAAGCACGGAGACGCGGTCGTGGGCCTGATGAAACTGGGCTTCTCGATCATCGAAGTGGGCTCGGTCACGCCGCAGCCGCAGCCCGGGAATCCCAAGCCGAGGGTCTTCCGACTCCCGGAGGACGGAGCCGTCATCAACAGATATGGCTTCAACAGCATAGGGCACGACGAAGTATACAAAAAGCTGGAAGGAATCGAGAAGGCCGTAATGAATCGGGCACTCCTCGGCGTGAATCTGGGAAAGAATAAGCTCTCCGATGATGCAGGGAAGGATTATGTGTCCGGCATTGAAAAGTTCTCGGATGTGGCTGATTATTTTGTTGTGAACGTTAGCAGCCCTAACACCCCCGGCCTGAGATCATTACAAAACAAGAACGAACTAGAAAACTTATTGACCGAAATAAACAGAGCTAGAAAACGGCGGAACTCGAACAAGCCGCTGCTCCTCAAACTGGCTCCGGACCTGAACGAGGACGAGCTAAAAGACGTGGTCAATGTCATTGGCAAACAGCACAGCAAGGTTGACGGCCTGATCATATCGAACACGACCGTCGAGAGGCAGAGCCTCAAGAATAAGGAGTTTGTGAACGAACCGGGCGGCCTGAGCGGGAAGCCGCTGACCAACAGATCTACGGAGATGATCAGGGACGTGTACAGATTAACTAAAGGTAAAGTTCCGATAATAGGAGTAGGGGGAGTGTTCACGGGGCGTGACGCGTATGAGAAGATCCTGGCGGGCGCCGGCGCCGTGCAAGTGTACACCGCGCTCATCTACCACGGACCTCCGGTCGTGAGGAGGATAAAGGACGAACTGGCCGAGCTACTAGAGAGGGACGGATATACTTCCGTCAATGACGCCGTCGGTAAAGGAGTTAAATAG
Protein
MSQKKDPLKKIRSLCYVTIGGAIAYQYIYYKKDFNSYYENVLQPLSQYLSPEVAHRIGVAAIKHGLFPPDQNEDPKILKTRLLDYDLSNPLGIAAGFDKHGDAVVGLMKLGFSIIEVGSVTPQPQPGNPKPRVFRLPEDGAVINRYGFNSIGHDEVYKKLEGIEKAVMNRALLGVNLGKNKLSDDAGKDYVSGIEKFSDVADYFVVNVSSPNTPGLRSLQNKNELENLLTEINRARKRRNSNKPLLLKLAPDLNEDELKDVVNVIGKQHSKVDGLIISNTTVERQSLKNKEFVNEPGGLSGKPLTNRSTEMIRDVYRLTKGKVPIIGVGGVFTGRDAYEKILAGAGAVQVYTALIYHGPPVVRRIKDELAELLERDGYTSVNDAVGKGVK
Summary
Catalytic Activity
(S)-dihydroorotate + a quinone = a quinol + orotate
Cofactor
FMN
Similarity
Belongs to the dihydroorotate dehydrogenase family. Type 2 subfamily.
Feature
chain Dihydroorotate dehydrogenase (quinone), mitochondrial
Uniprot
A0A2A4JRW4
A0A2W1BM13
H9JQT1
A0A212FE82
A0A0N0P9Y3
A0A194RR17
+ More
A0A2H1VTV9 A0A0L7KQR1 A0A232FK56 A0A151I8L9 K7IV72 A0A0L7QKU1 E9IG07 A0A0J7KDC7 A0A195FHK3 A0A158NY02 B0W4C3 F4X5R2 A0A195BQN8 A0A1Q3F595 A0A026VZ14 A0A154PUY4 D6X3Z9 A0A182VB65 A0A0C9QVV7 W5JPS2 A0A182U258 A0A182XB88 A0A182LH38 Q7PYN9 E2AD36 A0A1Y9H2S5 A0A182JX82 A0A182I031 A0A182GVP7 A0A2J7PNT2 A0A084VTW7 A0A182ILH6 A0A182PRI2 Q177S6 A0A1Y9HEN3 A0A182YAT0 A0A182T4P1 A0A182VZ37 A0A0K8TKL0 A0A182LS84 U5EXS8 A0A2A3ET49 A0A1B6G2N7 A0A0T6AUM4 A0A067R2K3 A0A1I8PEP4 L7LZA5 A0A088AF14 T1PH49 A0A1W4WUH8 A0A1I8MQ26 A0A1A9ZA90 A0A1J1J568 A0A1B6DRI4 E2BRB5 W8C685 A0A2A3EUN4 G3MQA6 E9GTU3 A0A1B6L4R0 A0A0A1XQP5 A0A3S0ZGA9 A0A0K8T8P9 A0A034V1M0 A0A146M731 A0A0A9WG38 A0A0K8URE4 A0A0P5YWH4 A0A0P6GRI0 A0A293MVJ8 A7SLG1 E0VQ82 A0A0P6AMA6 A0A1Y1NH83 A0A2P8XK77 A0A0V0GC74 A0A336M2C9 A0A224YL24 A0A0L0CPQ6 A0A0P5YQI2 A0A2R5L8K3 A0A0P6FX32 A0A2T7NGX8 A0A069DTM0 A0A1B0CTK1 A0A0P5HQ02 C3YFF0 A0A131YUV3 A0A0N8B0J4 A0A0P5HMR5 A0A0P5HKW6 A0A0N8EM82 A0A210PJJ5 A0A1A9VGV6
A0A2H1VTV9 A0A0L7KQR1 A0A232FK56 A0A151I8L9 K7IV72 A0A0L7QKU1 E9IG07 A0A0J7KDC7 A0A195FHK3 A0A158NY02 B0W4C3 F4X5R2 A0A195BQN8 A0A1Q3F595 A0A026VZ14 A0A154PUY4 D6X3Z9 A0A182VB65 A0A0C9QVV7 W5JPS2 A0A182U258 A0A182XB88 A0A182LH38 Q7PYN9 E2AD36 A0A1Y9H2S5 A0A182JX82 A0A182I031 A0A182GVP7 A0A2J7PNT2 A0A084VTW7 A0A182ILH6 A0A182PRI2 Q177S6 A0A1Y9HEN3 A0A182YAT0 A0A182T4P1 A0A182VZ37 A0A0K8TKL0 A0A182LS84 U5EXS8 A0A2A3ET49 A0A1B6G2N7 A0A0T6AUM4 A0A067R2K3 A0A1I8PEP4 L7LZA5 A0A088AF14 T1PH49 A0A1W4WUH8 A0A1I8MQ26 A0A1A9ZA90 A0A1J1J568 A0A1B6DRI4 E2BRB5 W8C685 A0A2A3EUN4 G3MQA6 E9GTU3 A0A1B6L4R0 A0A0A1XQP5 A0A3S0ZGA9 A0A0K8T8P9 A0A034V1M0 A0A146M731 A0A0A9WG38 A0A0K8URE4 A0A0P5YWH4 A0A0P6GRI0 A0A293MVJ8 A7SLG1 E0VQ82 A0A0P6AMA6 A0A1Y1NH83 A0A2P8XK77 A0A0V0GC74 A0A336M2C9 A0A224YL24 A0A0L0CPQ6 A0A0P5YQI2 A0A2R5L8K3 A0A0P6FX32 A0A2T7NGX8 A0A069DTM0 A0A1B0CTK1 A0A0P5HQ02 C3YFF0 A0A131YUV3 A0A0N8B0J4 A0A0P5HMR5 A0A0P5HKW6 A0A0N8EM82 A0A210PJJ5 A0A1A9VGV6
EC Number
1.3.5.2
Pubmed
28756777
19121390
22118469
26354079
26227816
28648823
+ More
20075255 21282665 21347285 21719571 24508170 30249741 18362917 19820115 20920257 23761445 20966253 12364791 14747013 17210077 20798317 26483478 24438588 17510324 25244985 26369729 24845553 25576852 25315136 24495485 22216098 21292972 25830018 25348373 26823975 25401762 17615350 20566863 28004739 29403074 28797301 26108605 26334808 18563158 26830274 28812685
20075255 21282665 21347285 21719571 24508170 30249741 18362917 19820115 20920257 23761445 20966253 12364791 14747013 17210077 20798317 26483478 24438588 17510324 25244985 26369729 24845553 25576852 25315136 24495485 22216098 21292972 25830018 25348373 26823975 25401762 17615350 20566863 28004739 29403074 28797301 26108605 26334808 18563158 26830274 28812685
EMBL
NWSH01000798
PCG74150.1
KZ150160
PZC72723.1
BABH01028744
AGBW02008970
+ More
OWR52030.1 KQ459037 KPJ04273.1 KQ459833 KPJ19839.1 ODYU01004426 SOQ44281.1 JTDY01006866 KOB65633.1 NNAY01000129 OXU30707.1 KQ978339 KYM95014.1 AAZX01005118 KQ414940 KOC59237.1 GL762905 EFZ20496.1 LBMM01009139 KMQ88362.1 KQ981606 KYN39484.1 ADTU01000548 DS231836 EDS33208.1 GL888740 EGI58213.1 KQ976423 KYM88886.1 GFDL01012319 JAV22726.1 KK107555 QOIP01000002 EZA49012.1 RLU26168.1 KQ435180 KZC15010.1 KQ971379 EEZ97344.2 GBYB01007884 JAG77651.1 ADMH02000517 ETN66131.1 AAAB01008987 EAA01242.5 GL438658 EFN68672.1 APCN01002042 JXUM01019119 KQ560533 KXJ81955.1 NEVH01023316 PNF17971.1 ATLV01016520 KE525092 KFB41411.1 CH477371 EAT42442.1 GDAI01002940 JAI14663.1 AXCM01001079 GANO01002394 JAB57477.1 KZ288185 PBC34925.1 GECZ01013072 JAS56697.1 LJIG01022760 KRT78863.1 KK852747 KDR17253.1 GACK01007929 JAA57105.1 KA648142 AFP62771.1 CVRI01000072 CRL07555.1 GEDC01009002 JAS28296.1 GL449942 EFN81778.1 GAMC01001267 JAC05289.1 PBC34926.1 JO844057 AEO35674.1 GL732564 EFX77146.1 GEBQ01021264 JAT18713.1 GBXI01001419 JAD12873.1 RQTK01000620 RUS76978.1 GBRD01004326 JAG61495.1 GAKP01021801 JAC37151.1 GDHC01004042 JAQ14587.1 GBHO01037238 JAG06366.1 GDHF01023067 JAI29247.1 GDIP01142101 GDIP01106142 GDIP01052358 LRGB01000868 JAL61613.1 JAM51357.1 KZS15672.1 GDIQ01029833 JAN64904.1 GFWV01019411 MAA44139.1 DS469698 EDO35448.1 DS235402 EEB15538.1 GDIP01027258 JAM76457.1 GEZM01003120 JAV96908.1 PYGN01001878 PSN32364.1 GECL01000756 JAP05368.1 UFQT01000446 SSX24404.1 GFPF01007201 MAA18347.1 JRES01000088 KNC34241.1 GDIP01055120 JAM48595.1 GGLE01001697 MBY05823.1 GDIQ01225911 GDIQ01052525 JAN42212.1 PZQS01000012 PVD20405.1 GBGD01001778 JAC87111.1 AJWK01027747 GDIQ01224708 JAK27017.1 GG666509 EEN60913.1 GEDV01005544 JAP83013.1 GDIQ01224707 JAK27018.1 GDIQ01225912 JAK25813.1 GDIQ01225910 JAK25815.1 GDIQ01013012 JAN81725.1 NEDP02076522 OWF36657.1
OWR52030.1 KQ459037 KPJ04273.1 KQ459833 KPJ19839.1 ODYU01004426 SOQ44281.1 JTDY01006866 KOB65633.1 NNAY01000129 OXU30707.1 KQ978339 KYM95014.1 AAZX01005118 KQ414940 KOC59237.1 GL762905 EFZ20496.1 LBMM01009139 KMQ88362.1 KQ981606 KYN39484.1 ADTU01000548 DS231836 EDS33208.1 GL888740 EGI58213.1 KQ976423 KYM88886.1 GFDL01012319 JAV22726.1 KK107555 QOIP01000002 EZA49012.1 RLU26168.1 KQ435180 KZC15010.1 KQ971379 EEZ97344.2 GBYB01007884 JAG77651.1 ADMH02000517 ETN66131.1 AAAB01008987 EAA01242.5 GL438658 EFN68672.1 APCN01002042 JXUM01019119 KQ560533 KXJ81955.1 NEVH01023316 PNF17971.1 ATLV01016520 KE525092 KFB41411.1 CH477371 EAT42442.1 GDAI01002940 JAI14663.1 AXCM01001079 GANO01002394 JAB57477.1 KZ288185 PBC34925.1 GECZ01013072 JAS56697.1 LJIG01022760 KRT78863.1 KK852747 KDR17253.1 GACK01007929 JAA57105.1 KA648142 AFP62771.1 CVRI01000072 CRL07555.1 GEDC01009002 JAS28296.1 GL449942 EFN81778.1 GAMC01001267 JAC05289.1 PBC34926.1 JO844057 AEO35674.1 GL732564 EFX77146.1 GEBQ01021264 JAT18713.1 GBXI01001419 JAD12873.1 RQTK01000620 RUS76978.1 GBRD01004326 JAG61495.1 GAKP01021801 JAC37151.1 GDHC01004042 JAQ14587.1 GBHO01037238 JAG06366.1 GDHF01023067 JAI29247.1 GDIP01142101 GDIP01106142 GDIP01052358 LRGB01000868 JAL61613.1 JAM51357.1 KZS15672.1 GDIQ01029833 JAN64904.1 GFWV01019411 MAA44139.1 DS469698 EDO35448.1 DS235402 EEB15538.1 GDIP01027258 JAM76457.1 GEZM01003120 JAV96908.1 PYGN01001878 PSN32364.1 GECL01000756 JAP05368.1 UFQT01000446 SSX24404.1 GFPF01007201 MAA18347.1 JRES01000088 KNC34241.1 GDIP01055120 JAM48595.1 GGLE01001697 MBY05823.1 GDIQ01225911 GDIQ01052525 JAN42212.1 PZQS01000012 PVD20405.1 GBGD01001778 JAC87111.1 AJWK01027747 GDIQ01224708 JAK27017.1 GG666509 EEN60913.1 GEDV01005544 JAP83013.1 GDIQ01224707 JAK27018.1 GDIQ01225912 JAK25813.1 GDIQ01225910 JAK25815.1 GDIQ01013012 JAN81725.1 NEDP02076522 OWF36657.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000215335 UP000078542 UP000002358 UP000053825 UP000036403 UP000078541 UP000005205 UP000002320 UP000007755 UP000078540 UP000053097 UP000279307 UP000076502 UP000007266 UP000075903 UP000000673 UP000075902 UP000076407 UP000075882 UP000007062 UP000000311 UP000075884 UP000075881 UP000075840 UP000069940 UP000249989 UP000235965 UP000030765 UP000075880 UP000075885 UP000008820 UP000075900 UP000076408 UP000075901 UP000075920 UP000075883 UP000242457 UP000027135 UP000095300 UP000005203 UP000192223 UP000095301 UP000092445 UP000183832 UP000008237 UP000000305 UP000271974 UP000076858 UP000001593 UP000009046 UP000245037 UP000037069 UP000245119 UP000092461 UP000001554 UP000242188 UP000078200
UP000215335 UP000078542 UP000002358 UP000053825 UP000036403 UP000078541 UP000005205 UP000002320 UP000007755 UP000078540 UP000053097 UP000279307 UP000076502 UP000007266 UP000075903 UP000000673 UP000075902 UP000076407 UP000075882 UP000007062 UP000000311 UP000075884 UP000075881 UP000075840 UP000069940 UP000249989 UP000235965 UP000030765 UP000075880 UP000075885 UP000008820 UP000075900 UP000076408 UP000075901 UP000075920 UP000075883 UP000242457 UP000027135 UP000095300 UP000005203 UP000192223 UP000095301 UP000092445 UP000183832 UP000008237 UP000000305 UP000271974 UP000076858 UP000001593 UP000009046 UP000245037 UP000037069 UP000245119 UP000092461 UP000001554 UP000242188 UP000078200
PRIDE
Interpro
IPR005719
Dihydroorotate_DH_2
+ More
IPR013785 Aldolase_TIM
IPR005720 Dihydroorotate_DH
IPR001295 Dihydroorotate_DH_CS
IPR012135 Dihydroorotate_DH_1_2
IPR005225 Small_GTP-bd_dom
IPR027417 P-loop_NTPase
IPR001806 Small_GTPase
IPR004117 7tm6_olfct_rcpt
IPR036691 Endo/exonu/phosph_ase_sf
IPR008636 Hook-like_fam
IPR036872 CH_dom_sf
IPR013154 ADH_N
IPR011032 GroES-like_sf
IPR013149 ADH_C
IPR036291 NAD(P)-bd_dom_sf
IPR013785 Aldolase_TIM
IPR005720 Dihydroorotate_DH
IPR001295 Dihydroorotate_DH_CS
IPR012135 Dihydroorotate_DH_1_2
IPR005225 Small_GTP-bd_dom
IPR027417 P-loop_NTPase
IPR001806 Small_GTPase
IPR004117 7tm6_olfct_rcpt
IPR036691 Endo/exonu/phosph_ase_sf
IPR008636 Hook-like_fam
IPR036872 CH_dom_sf
IPR013154 ADH_N
IPR011032 GroES-like_sf
IPR013149 ADH_C
IPR036291 NAD(P)-bd_dom_sf
Gene 3D
ProteinModelPortal
A0A2A4JRW4
A0A2W1BM13
H9JQT1
A0A212FE82
A0A0N0P9Y3
A0A194RR17
+ More
A0A2H1VTV9 A0A0L7KQR1 A0A232FK56 A0A151I8L9 K7IV72 A0A0L7QKU1 E9IG07 A0A0J7KDC7 A0A195FHK3 A0A158NY02 B0W4C3 F4X5R2 A0A195BQN8 A0A1Q3F595 A0A026VZ14 A0A154PUY4 D6X3Z9 A0A182VB65 A0A0C9QVV7 W5JPS2 A0A182U258 A0A182XB88 A0A182LH38 Q7PYN9 E2AD36 A0A1Y9H2S5 A0A182JX82 A0A182I031 A0A182GVP7 A0A2J7PNT2 A0A084VTW7 A0A182ILH6 A0A182PRI2 Q177S6 A0A1Y9HEN3 A0A182YAT0 A0A182T4P1 A0A182VZ37 A0A0K8TKL0 A0A182LS84 U5EXS8 A0A2A3ET49 A0A1B6G2N7 A0A0T6AUM4 A0A067R2K3 A0A1I8PEP4 L7LZA5 A0A088AF14 T1PH49 A0A1W4WUH8 A0A1I8MQ26 A0A1A9ZA90 A0A1J1J568 A0A1B6DRI4 E2BRB5 W8C685 A0A2A3EUN4 G3MQA6 E9GTU3 A0A1B6L4R0 A0A0A1XQP5 A0A3S0ZGA9 A0A0K8T8P9 A0A034V1M0 A0A146M731 A0A0A9WG38 A0A0K8URE4 A0A0P5YWH4 A0A0P6GRI0 A0A293MVJ8 A7SLG1 E0VQ82 A0A0P6AMA6 A0A1Y1NH83 A0A2P8XK77 A0A0V0GC74 A0A336M2C9 A0A224YL24 A0A0L0CPQ6 A0A0P5YQI2 A0A2R5L8K3 A0A0P6FX32 A0A2T7NGX8 A0A069DTM0 A0A1B0CTK1 A0A0P5HQ02 C3YFF0 A0A131YUV3 A0A0N8B0J4 A0A0P5HMR5 A0A0P5HKW6 A0A0N8EM82 A0A210PJJ5 A0A1A9VGV6
A0A2H1VTV9 A0A0L7KQR1 A0A232FK56 A0A151I8L9 K7IV72 A0A0L7QKU1 E9IG07 A0A0J7KDC7 A0A195FHK3 A0A158NY02 B0W4C3 F4X5R2 A0A195BQN8 A0A1Q3F595 A0A026VZ14 A0A154PUY4 D6X3Z9 A0A182VB65 A0A0C9QVV7 W5JPS2 A0A182U258 A0A182XB88 A0A182LH38 Q7PYN9 E2AD36 A0A1Y9H2S5 A0A182JX82 A0A182I031 A0A182GVP7 A0A2J7PNT2 A0A084VTW7 A0A182ILH6 A0A182PRI2 Q177S6 A0A1Y9HEN3 A0A182YAT0 A0A182T4P1 A0A182VZ37 A0A0K8TKL0 A0A182LS84 U5EXS8 A0A2A3ET49 A0A1B6G2N7 A0A0T6AUM4 A0A067R2K3 A0A1I8PEP4 L7LZA5 A0A088AF14 T1PH49 A0A1W4WUH8 A0A1I8MQ26 A0A1A9ZA90 A0A1J1J568 A0A1B6DRI4 E2BRB5 W8C685 A0A2A3EUN4 G3MQA6 E9GTU3 A0A1B6L4R0 A0A0A1XQP5 A0A3S0ZGA9 A0A0K8T8P9 A0A034V1M0 A0A146M731 A0A0A9WG38 A0A0K8URE4 A0A0P5YWH4 A0A0P6GRI0 A0A293MVJ8 A7SLG1 E0VQ82 A0A0P6AMA6 A0A1Y1NH83 A0A2P8XK77 A0A0V0GC74 A0A336M2C9 A0A224YL24 A0A0L0CPQ6 A0A0P5YQI2 A0A2R5L8K3 A0A0P6FX32 A0A2T7NGX8 A0A069DTM0 A0A1B0CTK1 A0A0P5HQ02 C3YFF0 A0A131YUV3 A0A0N8B0J4 A0A0P5HMR5 A0A0P5HKW6 A0A0N8EM82 A0A210PJJ5 A0A1A9VGV6
PDB
6FMD
E-value=1.00955e-85,
Score=807
Ontologies
PATHWAY
GO
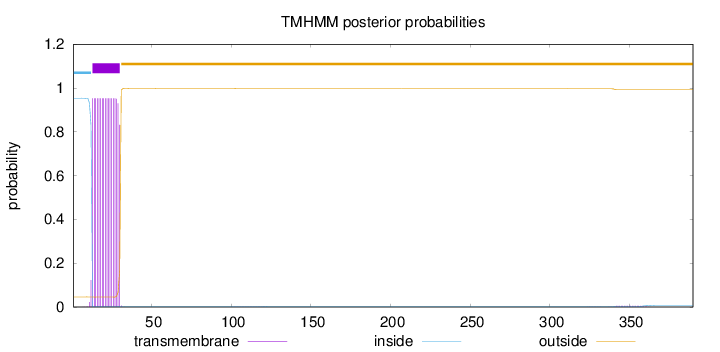
Topology
Subcellular location
Mitochondrion inner membrane
Length:
390
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.2842199999999
Exp number, first 60 AAs:
17.1666
Total prob of N-in:
0.95388
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 30
outside
31 - 390
Population Genetic Test Statistics
Pi
188.257988
Theta
165.881556
Tajima's D
0.328469
CLR
0.0228
CSRT
0.464176791160442
Interpretation
Uncertain
