Gene
KWMTBOMO06313
Pre Gene Modal
BGIBMGA011886
Annotation
PREDICTED:_peroxidase-like_isoform_X1_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 2.335
Sequence
CDS
ATGAAAGGCAGAGACCTAGGTTTACCGCCCTACAACGATTATCGTCGGCTGTGCGGTCTGCCAGTCGCGAAGAAGTTTAAAGATTTGCTACGTTGGATGAGAAGTGAGCAAGTTGAAGCGATGTCGCTTCAGTATGAGAGCACGGAGGACATTGACCTAATGGCTGGTATAATAGCGGAGTGGCCAGCTAAAGACGCGGTGGTTGGACCGACGCTGCGCTGTATCATGGCAACACAGATGAAAAGATGGCGGCGCGCCGACAGGTTCTGGTATGAGAACGACATACATCCGGGTGCTTTAACCCAAGATCAATTGGTGGAAATAAGGAAAATATCTATGGCCAAAATTATGTGCGACCACGGGGATCAAGTTGAAGATATACAACCAAATGTGTTCATCTTACCTGGATACGGGTAA
Protein
MKGRDLGLPPYNDYRRLCGLPVAKKFKDLLRWMRSEQVEAMSLQYESTEDIDLMAGIIAEWPAKDAVVGPTLRCIMATQMKRWRRADRFWYENDIHPGALTQDQLVEIRKISMAKIMCDHGDQVEDIQPNVFILPGYG
Summary
Uniprot
H9JQT0
A0A0N0PED8
A0A3S2L850
A0A3S2LP82
A0A194Q3N0
A0A1W0XF69
+ More
D6WB23 A0A0V1DFW7 A0A0V1DG33 A0A139WNI1 A0A3P6TXW3 A0A0V0UCN0 A0A0V0SM66 A0A0V1LQ01 A0A0V0VF55 A0A0V0VER0 A0A0V0UCL7 A0A0V0VEP3 A0A0V0VEM5 A0A0V0SMI6 A0A1D1UVK4 A0A3P7K3P4 A0A0V1NA40 A0A0V1I8Q1 A0A0V1I8M9 A0A0V1N9Q9 A0A0V1I8N2 A0A0V1C1H7 A0A0V0WR39 A0A0V1A116 A0A0V1EJR1 A0A0V1EJ09 A0A0V1EJE8 A0A0V1JFF6 A0A0V1FT12 A0A0V1EIN1 A0A0V1EIN4 A0A0V1JRK2 A0A0V1JRK7 A0A0D6LK20 A0A0V1PPD5 A0A2A2JDR5 A0A2A2L2J1 A0A016TGN8 A0A0N0BI50 A0A016THP1 J9EG18 A0A368FXY0 A0A1B6IGX2 A0A0J7KKC2 A0A016WZC2 A0A3L8DTP5 A0A1Y3APM9 A0A1B6C8P6 A0A0I9N7J2 A0A026W3J3 E2BCE4 A0A088AE08 A0A195FC13 A0A3P7EAB6 A0A195BAD8 A0A2G9UAZ1 E1G4R9 A0A0N4UUD4 A0A1I8C365 A0A158NBE1 A0A0N4TR06 A0A077Z659 A0A3P7DE81 A0A1I8EHT1 A0A0K0J7G2 A0A0L7LKV1 A0A183I2S7 A0A1B6MCZ0 V9IAE9 E2ADZ2 A0A0C9R888 A0A2A3E974 A0A1B6CXE7 A0A2A4IUP3 V9ICS7 F4X8U9 E9IRB0 A0A2S2NJQ9 A0A151X152 Q18647 A0A238BS93 A0A2A2KER3 T1IJ78 A0A1D2NGG8 A0A1I7VJ16 A0A2H1UZX5 A0A1B6DWT2 A0A085NFK7 A0A3P6TJ72 E0VJ44 A0A368GRF7 A0A2A2JE10 A0A0M3IZG9 A0A261AGS1
D6WB23 A0A0V1DFW7 A0A0V1DG33 A0A139WNI1 A0A3P6TXW3 A0A0V0UCN0 A0A0V0SM66 A0A0V1LQ01 A0A0V0VF55 A0A0V0VER0 A0A0V0UCL7 A0A0V0VEP3 A0A0V0VEM5 A0A0V0SMI6 A0A1D1UVK4 A0A3P7K3P4 A0A0V1NA40 A0A0V1I8Q1 A0A0V1I8M9 A0A0V1N9Q9 A0A0V1I8N2 A0A0V1C1H7 A0A0V0WR39 A0A0V1A116 A0A0V1EJR1 A0A0V1EJ09 A0A0V1EJE8 A0A0V1JFF6 A0A0V1FT12 A0A0V1EIN1 A0A0V1EIN4 A0A0V1JRK2 A0A0V1JRK7 A0A0D6LK20 A0A0V1PPD5 A0A2A2JDR5 A0A2A2L2J1 A0A016TGN8 A0A0N0BI50 A0A016THP1 J9EG18 A0A368FXY0 A0A1B6IGX2 A0A0J7KKC2 A0A016WZC2 A0A3L8DTP5 A0A1Y3APM9 A0A1B6C8P6 A0A0I9N7J2 A0A026W3J3 E2BCE4 A0A088AE08 A0A195FC13 A0A3P7EAB6 A0A195BAD8 A0A2G9UAZ1 E1G4R9 A0A0N4UUD4 A0A1I8C365 A0A158NBE1 A0A0N4TR06 A0A077Z659 A0A3P7DE81 A0A1I8EHT1 A0A0K0J7G2 A0A0L7LKV1 A0A183I2S7 A0A1B6MCZ0 V9IAE9 E2ADZ2 A0A0C9R888 A0A2A3E974 A0A1B6CXE7 A0A2A4IUP3 V9ICS7 F4X8U9 E9IRB0 A0A2S2NJQ9 A0A151X152 Q18647 A0A238BS93 A0A2A2KER3 T1IJ78 A0A1D2NGG8 A0A1I7VJ16 A0A2H1UZX5 A0A1B6DWT2 A0A085NFK7 A0A3P6TJ72 E0VJ44 A0A368GRF7 A0A2A2JE10 A0A0M3IZG9 A0A261AGS1
Pubmed
EMBL
BABH01028746
BABH01028747
BABH01028748
KQ459837
KPJ19761.1
RSAL01000844
+ More
RVE41111.1 RSAL01001640 RVE40745.1 KQ459564 KPI99614.1 MTYJ01000001 OQV26124.1 KQ971311 EEZ99334.2 JYDI01000005 KRY60541.1 KRY60542.1 KYB29427.1 UYRX01000802 VDK86345.1 JYDJ01000022 KRX48984.1 JYDL01000002 KRX27844.1 JYDW01000017 KRZ61524.1 JYDN01000042 KRX61985.1 KRX61984.1 KRX48986.1 KRX61986.1 KRX61982.1 KRX27846.1 BDGG01000002 GAU92485.1 UYYB01120607 VDM83017.1 JYDO01000001 KRZ80716.1 JYDP01000001 KRZ19194.1 KRZ19196.1 KRZ80717.1 KRZ19195.1 JYDH01000002 KRY43094.1 JYDK01000071 KRX78121.1 JYDQ01000044 KRY18614.1 JYDR01000032 KRY73682.1 KRY73676.1 KRY73681.1 JYDS01000007 KRZ33746.1 JYDT01000035 KRY89061.1 KRY73684.1 KRY73677.1 JYDV01000056 KRZ37626.1 KRZ37625.1 KE125055 EPB72289.1 JYDM01000001 KRZ98104.1 LIAE01010510 PAV59662.1 LIAE01007263 PAV80481.1 JARK01001438 EYC02099.1 KQ435733 KOX77231.1 EYC02098.1 ADBV01003944 EJW81073.1 JOJR01000502 RCN37044.1 GECU01021534 JAS86172.1 LBMM01006170 KMQ90863.1 JARK01000056 EYC44587.1 QOIP01000004 RLU23754.1 MUJZ01068849 OTF69784.1 GEDC01027673 JAS09625.1 LN854926 CTP80702.1 KK107453 EZA50655.1 GL447257 EFN86680.1 KQ981673 KYN38170.1 UYWW01004431 VDM13507.1 KQ976540 KYM81185.1 KZ347648 PIO67353.1 JH712764 EFO20336.2 UXUI01002462 VDD85556.1 ADTU01010985 ADTU01010986 ADTU01010987 UZAD01013209 VDN92204.1 HG805947 CDW55319.1 UYWW01000149 VDM07493.1 JTDY01000732 KOB76072.1 UZAJ01040554 VDP15339.1 GEBQ01006176 JAT33801.1 JR038044 AEY58093.1 GL438827 EFN68225.1 GBYB01004245 JAG74012.1 KZ288315 PBC28333.1 GEDC01019210 GEDC01003551 JAS18088.1 JAS33747.1 NWSH01006414 PCG63451.1 JR038045 AEY58094.1 GL888932 EGI57368.1 GL765107 EFZ16876.1 GGMR01004804 MBY17423.1 KQ982603 KYQ53986.1 BX284604 CCD66594.1 KZ270029 OZC07550.1 LIAE01008806 PAV72368.1 AFFK01014481 LJIJ01000056 ODN04056.1 ODYU01000046 SOQ34155.1 GEDC01007178 JAS30120.1 KL367506 KFD68253.1 UYRX01000675 VDK85217.1 DS235219 EEB13400.1 JOJR01000070 RCN46952.1 LIAE01010491 PAV59943.1 UYRR01000388 VDK17833.1 NMWX01000006 OZF97313.1
RVE41111.1 RSAL01001640 RVE40745.1 KQ459564 KPI99614.1 MTYJ01000001 OQV26124.1 KQ971311 EEZ99334.2 JYDI01000005 KRY60541.1 KRY60542.1 KYB29427.1 UYRX01000802 VDK86345.1 JYDJ01000022 KRX48984.1 JYDL01000002 KRX27844.1 JYDW01000017 KRZ61524.1 JYDN01000042 KRX61985.1 KRX61984.1 KRX48986.1 KRX61986.1 KRX61982.1 KRX27846.1 BDGG01000002 GAU92485.1 UYYB01120607 VDM83017.1 JYDO01000001 KRZ80716.1 JYDP01000001 KRZ19194.1 KRZ19196.1 KRZ80717.1 KRZ19195.1 JYDH01000002 KRY43094.1 JYDK01000071 KRX78121.1 JYDQ01000044 KRY18614.1 JYDR01000032 KRY73682.1 KRY73676.1 KRY73681.1 JYDS01000007 KRZ33746.1 JYDT01000035 KRY89061.1 KRY73684.1 KRY73677.1 JYDV01000056 KRZ37626.1 KRZ37625.1 KE125055 EPB72289.1 JYDM01000001 KRZ98104.1 LIAE01010510 PAV59662.1 LIAE01007263 PAV80481.1 JARK01001438 EYC02099.1 KQ435733 KOX77231.1 EYC02098.1 ADBV01003944 EJW81073.1 JOJR01000502 RCN37044.1 GECU01021534 JAS86172.1 LBMM01006170 KMQ90863.1 JARK01000056 EYC44587.1 QOIP01000004 RLU23754.1 MUJZ01068849 OTF69784.1 GEDC01027673 JAS09625.1 LN854926 CTP80702.1 KK107453 EZA50655.1 GL447257 EFN86680.1 KQ981673 KYN38170.1 UYWW01004431 VDM13507.1 KQ976540 KYM81185.1 KZ347648 PIO67353.1 JH712764 EFO20336.2 UXUI01002462 VDD85556.1 ADTU01010985 ADTU01010986 ADTU01010987 UZAD01013209 VDN92204.1 HG805947 CDW55319.1 UYWW01000149 VDM07493.1 JTDY01000732 KOB76072.1 UZAJ01040554 VDP15339.1 GEBQ01006176 JAT33801.1 JR038044 AEY58093.1 GL438827 EFN68225.1 GBYB01004245 JAG74012.1 KZ288315 PBC28333.1 GEDC01019210 GEDC01003551 JAS18088.1 JAS33747.1 NWSH01006414 PCG63451.1 JR038045 AEY58094.1 GL888932 EGI57368.1 GL765107 EFZ16876.1 GGMR01004804 MBY17423.1 KQ982603 KYQ53986.1 BX284604 CCD66594.1 KZ270029 OZC07550.1 LIAE01008806 PAV72368.1 AFFK01014481 LJIJ01000056 ODN04056.1 ODYU01000046 SOQ34155.1 GEDC01007178 JAS30120.1 KL367506 KFD68253.1 UYRX01000675 VDK85217.1 DS235219 EEB13400.1 JOJR01000070 RCN46952.1 LIAE01010491 PAV59943.1 UYRR01000388 VDK17833.1 NMWX01000006 OZF97313.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000053268
UP000007266
UP000054653
+ More
UP000277928 UP000055048 UP000054630 UP000054721 UP000054681 UP000186922 UP000054843 UP000055024 UP000054776 UP000054673 UP000054783 UP000054632 UP000054805 UP000054995 UP000054826 UP000054924 UP000218231 UP000024635 UP000053105 UP000004810 UP000252519 UP000036403 UP000279307 UP000053097 UP000008237 UP000005203 UP000078541 UP000270924 UP000078540 UP000038041 UP000274131 UP000095286 UP000005205 UP000038020 UP000278627 UP000030665 UP000093561 UP000006672 UP000037510 UP000050787 UP000000311 UP000242457 UP000218220 UP000007755 UP000075809 UP000001940 UP000094527 UP000095285 UP000009046 UP000036680 UP000216624
UP000277928 UP000055048 UP000054630 UP000054721 UP000054681 UP000186922 UP000054843 UP000055024 UP000054776 UP000054673 UP000054783 UP000054632 UP000054805 UP000054995 UP000054826 UP000054924 UP000218231 UP000024635 UP000053105 UP000004810 UP000252519 UP000036403 UP000279307 UP000053097 UP000008237 UP000005203 UP000078541 UP000270924 UP000078540 UP000038041 UP000274131 UP000095286 UP000005205 UP000038020 UP000278627 UP000030665 UP000093561 UP000006672 UP000037510 UP000050787 UP000000311 UP000242457 UP000218220 UP000007755 UP000075809 UP000001940 UP000094527 UP000095285 UP000009046 UP000036680 UP000216624
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JQT0
A0A0N0PED8
A0A3S2L850
A0A3S2LP82
A0A194Q3N0
A0A1W0XF69
+ More
D6WB23 A0A0V1DFW7 A0A0V1DG33 A0A139WNI1 A0A3P6TXW3 A0A0V0UCN0 A0A0V0SM66 A0A0V1LQ01 A0A0V0VF55 A0A0V0VER0 A0A0V0UCL7 A0A0V0VEP3 A0A0V0VEM5 A0A0V0SMI6 A0A1D1UVK4 A0A3P7K3P4 A0A0V1NA40 A0A0V1I8Q1 A0A0V1I8M9 A0A0V1N9Q9 A0A0V1I8N2 A0A0V1C1H7 A0A0V0WR39 A0A0V1A116 A0A0V1EJR1 A0A0V1EJ09 A0A0V1EJE8 A0A0V1JFF6 A0A0V1FT12 A0A0V1EIN1 A0A0V1EIN4 A0A0V1JRK2 A0A0V1JRK7 A0A0D6LK20 A0A0V1PPD5 A0A2A2JDR5 A0A2A2L2J1 A0A016TGN8 A0A0N0BI50 A0A016THP1 J9EG18 A0A368FXY0 A0A1B6IGX2 A0A0J7KKC2 A0A016WZC2 A0A3L8DTP5 A0A1Y3APM9 A0A1B6C8P6 A0A0I9N7J2 A0A026W3J3 E2BCE4 A0A088AE08 A0A195FC13 A0A3P7EAB6 A0A195BAD8 A0A2G9UAZ1 E1G4R9 A0A0N4UUD4 A0A1I8C365 A0A158NBE1 A0A0N4TR06 A0A077Z659 A0A3P7DE81 A0A1I8EHT1 A0A0K0J7G2 A0A0L7LKV1 A0A183I2S7 A0A1B6MCZ0 V9IAE9 E2ADZ2 A0A0C9R888 A0A2A3E974 A0A1B6CXE7 A0A2A4IUP3 V9ICS7 F4X8U9 E9IRB0 A0A2S2NJQ9 A0A151X152 Q18647 A0A238BS93 A0A2A2KER3 T1IJ78 A0A1D2NGG8 A0A1I7VJ16 A0A2H1UZX5 A0A1B6DWT2 A0A085NFK7 A0A3P6TJ72 E0VJ44 A0A368GRF7 A0A2A2JE10 A0A0M3IZG9 A0A261AGS1
D6WB23 A0A0V1DFW7 A0A0V1DG33 A0A139WNI1 A0A3P6TXW3 A0A0V0UCN0 A0A0V0SM66 A0A0V1LQ01 A0A0V0VF55 A0A0V0VER0 A0A0V0UCL7 A0A0V0VEP3 A0A0V0VEM5 A0A0V0SMI6 A0A1D1UVK4 A0A3P7K3P4 A0A0V1NA40 A0A0V1I8Q1 A0A0V1I8M9 A0A0V1N9Q9 A0A0V1I8N2 A0A0V1C1H7 A0A0V0WR39 A0A0V1A116 A0A0V1EJR1 A0A0V1EJ09 A0A0V1EJE8 A0A0V1JFF6 A0A0V1FT12 A0A0V1EIN1 A0A0V1EIN4 A0A0V1JRK2 A0A0V1JRK7 A0A0D6LK20 A0A0V1PPD5 A0A2A2JDR5 A0A2A2L2J1 A0A016TGN8 A0A0N0BI50 A0A016THP1 J9EG18 A0A368FXY0 A0A1B6IGX2 A0A0J7KKC2 A0A016WZC2 A0A3L8DTP5 A0A1Y3APM9 A0A1B6C8P6 A0A0I9N7J2 A0A026W3J3 E2BCE4 A0A088AE08 A0A195FC13 A0A3P7EAB6 A0A195BAD8 A0A2G9UAZ1 E1G4R9 A0A0N4UUD4 A0A1I8C365 A0A158NBE1 A0A0N4TR06 A0A077Z659 A0A3P7DE81 A0A1I8EHT1 A0A0K0J7G2 A0A0L7LKV1 A0A183I2S7 A0A1B6MCZ0 V9IAE9 E2ADZ2 A0A0C9R888 A0A2A3E974 A0A1B6CXE7 A0A2A4IUP3 V9ICS7 F4X8U9 E9IRB0 A0A2S2NJQ9 A0A151X152 Q18647 A0A238BS93 A0A2A2KER3 T1IJ78 A0A1D2NGG8 A0A1I7VJ16 A0A2H1UZX5 A0A1B6DWT2 A0A085NFK7 A0A3P6TJ72 E0VJ44 A0A368GRF7 A0A2A2JE10 A0A0M3IZG9 A0A261AGS1
PDB
5WDJ
E-value=2.47883e-17,
Score=210
Ontologies
GO
PANTHER
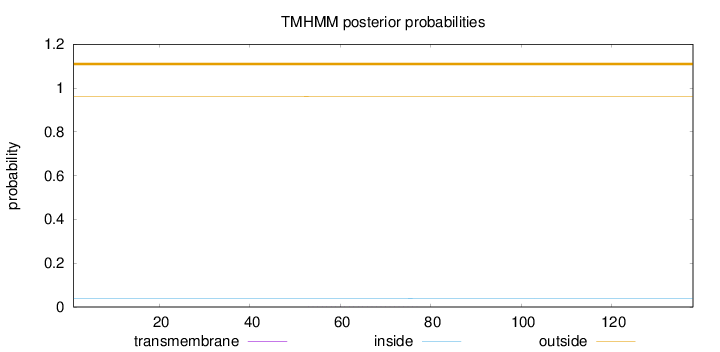
Topology
Length:
138
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02276
Exp number, first 60 AAs:
0.00686
Total prob of N-in:
0.03791
outside
1 - 138
Population Genetic Test Statistics
Pi
172.632222
Theta
133.16093
Tajima's D
1.184584
CLR
0.072757
CSRT
0.705414729263537
Interpretation
Uncertain
