Gene
KWMTBOMO06312
Pre Gene Modal
BGIBMGA011886
Annotation
PREDICTED:_peroxidase-like_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.14
Sequence
CDS
ATGGTGACCAACTACAACGTTTTCATCGCCGGAGACGTGACTTCAGTTCACGACACAGTGAACTATGTGGCTGTCACGACAGCTTGCTGCTCACCAGGTGGAGAGGCAGACCCGAGATGCCTACCGATCAGAGTGCCCGATGATGATGTGCATCTTCGTAGAAGCAACGTGAGATGTCTGAATTTGACACGTGCCATCACGTATCAGCGTCTGGGCTGTGTATCCAATAATGTGCCTGGAGAACGGATCAACACAGCAACGCCCATGCTCGATCTCTCGATCGTGTACGGCAACAACGCTATGGCTGCAGCCCGAGGAAGGCAGTACGAGGGTGGTAGACTCCGTGCTGACATATTGAAAGGCAAGGACTGGCCACCTTCAGGGTCGGCATTCTGCATCAACAACGAAGCCAATGAGACAAGATGCCACGACGCTCCGACGAACGGCGCCAATTCGCTTTTGGGCATCAACATGTTGTTCCTCTGGCTGTACCGGAACCACAATTCTATAGCCAGACGGCTTCAGACCCTGAACCGATGCTGGGACGACGATAGAATCTTCTTCACCGCTAAAGAAATCAACATAGCCATGTATTTGCAAATCCTGTACTATGAACTCATGCCTGTAGTTTTAGGAAAAGAATTTCTACTGAACAACAAAGTTATCTTTGACGTTTTGGGTCACGTCAATGACTATGACGATCGTCTAATGCCCACTGTCAGTCTGGAGTACATTATTGGAACTAGATGGTTCCACACTATACAGGAAGGGAGGCTGAGATTGTACACAAAAGAAGGGAAGATATACGATGAACAGCCAATAGTCGATTTGACTCTTCGGACTGGTGCATTACCCATTAATAATACCTTGGAGGGCGTAACCCAGGGGGCCTTCAGACAGCCGTGTGCCGACACCGATAATCTGGTAGATCCTGATGTAAGTTATACCGGTAGCTAA
Protein
MVTNYNVFIAGDVTSVHDTVNYVAVTTACCSPGGEADPRCLPIRVPDDDVHLRRSNVRCLNLTRAITYQRLGCVSNNVPGERINTATPMLDLSIVYGNNAMAAARGRQYEGGRLRADILKGKDWPPSGSAFCINNEANETRCHDAPTNGANSLLGINMLFLWLYRNHNSIARRLQTLNRCWDDDRIFFTAKEINIAMYLQILYYELMPVVLGKEFLLNNKVIFDVLGHVNDYDDRLMPTVSLEYIIGTRWFHTIQEGRLRLYTKEGKIYDEQPIVDLTLRTGALPINNTLEGVTQGAFRQPCADTDNLVDPDVSYTGS
Summary
Uniprot
H9JQT0
A0A3S2NMV9
A0A0N0PED8
A0A3S2L850
A0A194Q3N0
A0A2H1W9U4
+ More
A0A3S2NLC3 A0A2H1WAX3 A0A194Q7Y0 A0A2A4IUC0 A0A194RBC8 A0A2A4J7D3 A0A194Q9H2 A0A194QDV6 A0A2W1BYE5 I4DJ10 A0A194RAP6 A0A212ETD1 A0A2W1BT67 A0A2A4IUP3 A0A2W1BQ93 A0A2A4JM77 A0A2A4JMB0 H9ISL1 H9JT77 A0A2W1BXH1 A0A2W1BS42 A0A2H1WBG2 A0A2A4IUD5 A0A2H1WGR9 A0A2A4JI21 A0A088MGW5 A0A2H1W8U7 A0A2W1BT77 H9JT78 A0A2H1VUX0 H9IT78 A0A3S2M0V1 A0A1W4WMG7 E2BCE2 N6UAQ6 D6WCM3 A0A088SIJ3 A0A194PXJ3 A0A1B6MDD3 A0A3L8DTW7 A0A1B6DIG5 A0A1B6CUT2 A0A026W3J3 A0A1B6F7C3 E2B078 A0A3S2M8S7 A0A154PJR6 A0A0L7R2K3 A0A1Y9HDW6 A0A232F2X4 A0A1Y9IWB3 A0A194RNM9 A0A182Q218 Q7QDZ3 A0A182KV49 A0A182WXU8 A0A182VL61 A0A182TM70 A0A182I5W9 K7IYZ3 A0A182PRL2 A0A310SND6 A0A182MPT8 A0A067RJX3 A0A1Y9G8K4 A0A1Y9H2I7 A0A088AE09 A0A194PVX4 A0A0P4VM15 A0A2J7RAM0 A0A182KBY5 B0WM28 B0XE84 A0A084V9X6 A0A182RQS6 W5JNS3 A0A084WCK1 A0A182IZD9 B4M5J8 T1I7W3 A0A2A3EU99 A0A0L0CRA6 A0A182Y6T5 W5JF37 A0A1Y9HES0 A0A182K431 A0A182FTE8 A0A1A9W584
A0A3S2NLC3 A0A2H1WAX3 A0A194Q7Y0 A0A2A4IUC0 A0A194RBC8 A0A2A4J7D3 A0A194Q9H2 A0A194QDV6 A0A2W1BYE5 I4DJ10 A0A194RAP6 A0A212ETD1 A0A2W1BT67 A0A2A4IUP3 A0A2W1BQ93 A0A2A4JM77 A0A2A4JMB0 H9ISL1 H9JT77 A0A2W1BXH1 A0A2W1BS42 A0A2H1WBG2 A0A2A4IUD5 A0A2H1WGR9 A0A2A4JI21 A0A088MGW5 A0A2H1W8U7 A0A2W1BT77 H9JT78 A0A2H1VUX0 H9IT78 A0A3S2M0V1 A0A1W4WMG7 E2BCE2 N6UAQ6 D6WCM3 A0A088SIJ3 A0A194PXJ3 A0A1B6MDD3 A0A3L8DTW7 A0A1B6DIG5 A0A1B6CUT2 A0A026W3J3 A0A1B6F7C3 E2B078 A0A3S2M8S7 A0A154PJR6 A0A0L7R2K3 A0A1Y9HDW6 A0A232F2X4 A0A1Y9IWB3 A0A194RNM9 A0A182Q218 Q7QDZ3 A0A182KV49 A0A182WXU8 A0A182VL61 A0A182TM70 A0A182I5W9 K7IYZ3 A0A182PRL2 A0A310SND6 A0A182MPT8 A0A067RJX3 A0A1Y9G8K4 A0A1Y9H2I7 A0A088AE09 A0A194PVX4 A0A0P4VM15 A0A2J7RAM0 A0A182KBY5 B0WM28 B0XE84 A0A084V9X6 A0A182RQS6 W5JNS3 A0A084WCK1 A0A182IZD9 B4M5J8 T1I7W3 A0A2A3EU99 A0A0L0CRA6 A0A182Y6T5 W5JF37 A0A1Y9HES0 A0A182K431 A0A182FTE8 A0A1A9W584
Pubmed
EMBL
BABH01028746
BABH01028747
BABH01028748
RSAL01000231
RVE43837.1
KQ459837
+ More
KPJ19761.1 RSAL01000844 RVE41111.1 KQ459564 KPI99614.1 ODYU01007226 SOQ49823.1 RSAL01000046 RVE50539.1 ODYU01007325 SOQ50002.1 KQ459324 KPJ01647.1 NWSH01006580 PCG63355.1 KQ460416 KPJ14922.1 NWSH01002854 PCG67434.1 KPJ01645.1 KPJ01646.1 KZ149907 PZC78257.1 AK401278 BAM17900.1 KPJ14923.1 AGBW02012596 OWR44740.1 PZC78258.1 NWSH01006414 PCG63451.1 KZ149956 PZC76461.1 NWSH01001116 PCG72533.1 PCG72532.1 BABH01013455 BABH01034743 BABH01034744 PZC76463.1 PZC76464.1 ODYU01007479 SOQ50286.1 NWSH01006200 PCG63597.1 ODYU01008556 SOQ52260.1 NWSH01001320 PCG71707.1 KJ995811 AIN39494.1 ODYU01007063 SOQ49501.1 PZC78259.1 BABH01034746 BABH01034747 ODYU01004596 SOQ44641.1 BABH01013458 BABH01013459 BABH01013460 RSAL01000083 RVE48485.1 GL447257 EFN86678.1 APGK01042097 APGK01042098 KB741002 KB631984 ENN75712.1 ERL87705.1 KQ971317 EEZ99172.1 KM384736 AIO11227.1 KQ459589 KPI97479.1 GEBQ01006041 JAT33936.1 QOIP01000004 RLU23752.1 GEDC01011809 JAS25489.1 GEDC01020121 JAS17177.1 KK107453 EZA50655.1 GECZ01023621 JAS46148.1 GL444420 EFN60907.1 RSAL01000011 RVE53586.1 KQ434938 KZC12037.1 KQ414666 KOC65001.1 NNAY01001189 OXU24803.1 KQ459896 KPJ19443.1 AXCN02001762 AAAB01008848 EAA07042.4 APCN01003535 KQ760645 OAD59676.1 AXCM01004244 KK852427 KDR24107.1 KPI97477.1 GDKW01003332 JAI53263.1 NEVH01006567 PNF37876.1 DS231994 EDS30856.1 DS232811 EDS45917.1 ATLV01000829 ATLV01000830 KE523910 KFB34770.1 ADMH02000540 ETN66032.1 ATLV01022681 KE525335 KFB47945.1 CH940652 EDW58924.2 ACPB03011632 KZ288187 PBC34719.1 JRES01000032 KNC34772.1 ADMH02001590 ETN61943.1
KPJ19761.1 RSAL01000844 RVE41111.1 KQ459564 KPI99614.1 ODYU01007226 SOQ49823.1 RSAL01000046 RVE50539.1 ODYU01007325 SOQ50002.1 KQ459324 KPJ01647.1 NWSH01006580 PCG63355.1 KQ460416 KPJ14922.1 NWSH01002854 PCG67434.1 KPJ01645.1 KPJ01646.1 KZ149907 PZC78257.1 AK401278 BAM17900.1 KPJ14923.1 AGBW02012596 OWR44740.1 PZC78258.1 NWSH01006414 PCG63451.1 KZ149956 PZC76461.1 NWSH01001116 PCG72533.1 PCG72532.1 BABH01013455 BABH01034743 BABH01034744 PZC76463.1 PZC76464.1 ODYU01007479 SOQ50286.1 NWSH01006200 PCG63597.1 ODYU01008556 SOQ52260.1 NWSH01001320 PCG71707.1 KJ995811 AIN39494.1 ODYU01007063 SOQ49501.1 PZC78259.1 BABH01034746 BABH01034747 ODYU01004596 SOQ44641.1 BABH01013458 BABH01013459 BABH01013460 RSAL01000083 RVE48485.1 GL447257 EFN86678.1 APGK01042097 APGK01042098 KB741002 KB631984 ENN75712.1 ERL87705.1 KQ971317 EEZ99172.1 KM384736 AIO11227.1 KQ459589 KPI97479.1 GEBQ01006041 JAT33936.1 QOIP01000004 RLU23752.1 GEDC01011809 JAS25489.1 GEDC01020121 JAS17177.1 KK107453 EZA50655.1 GECZ01023621 JAS46148.1 GL444420 EFN60907.1 RSAL01000011 RVE53586.1 KQ434938 KZC12037.1 KQ414666 KOC65001.1 NNAY01001189 OXU24803.1 KQ459896 KPJ19443.1 AXCN02001762 AAAB01008848 EAA07042.4 APCN01003535 KQ760645 OAD59676.1 AXCM01004244 KK852427 KDR24107.1 KPI97477.1 GDKW01003332 JAI53263.1 NEVH01006567 PNF37876.1 DS231994 EDS30856.1 DS232811 EDS45917.1 ATLV01000829 ATLV01000830 KE523910 KFB34770.1 ADMH02000540 ETN66032.1 ATLV01022681 KE525335 KFB47945.1 CH940652 EDW58924.2 ACPB03011632 KZ288187 PBC34719.1 JRES01000032 KNC34772.1 ADMH02001590 ETN61943.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000218220
UP000007151
+ More
UP000192223 UP000008237 UP000019118 UP000030742 UP000007266 UP000279307 UP000053097 UP000000311 UP000076502 UP000053825 UP000075900 UP000215335 UP000075920 UP000075886 UP000007062 UP000075882 UP000076407 UP000075903 UP000075902 UP000075840 UP000002358 UP000075885 UP000075883 UP000027135 UP000069272 UP000075884 UP000005203 UP000235965 UP000075881 UP000002320 UP000030765 UP000000673 UP000075880 UP000008792 UP000015103 UP000242457 UP000037069 UP000076408 UP000091820
UP000192223 UP000008237 UP000019118 UP000030742 UP000007266 UP000279307 UP000053097 UP000000311 UP000076502 UP000053825 UP000075900 UP000215335 UP000075920 UP000075886 UP000007062 UP000075882 UP000076407 UP000075903 UP000075902 UP000075840 UP000002358 UP000075885 UP000075883 UP000027135 UP000069272 UP000075884 UP000005203 UP000235965 UP000075881 UP000002320 UP000030765 UP000000673 UP000075880 UP000008792 UP000015103 UP000242457 UP000037069 UP000076408 UP000091820
Interpro
IPR029585
Pxd-like
+ More
IPR037120 Haem_peroxidase_sf_animal
IPR019791 Haem_peroxidase_animal
IPR010255 Haem_peroxidase_sf
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR012337 RNaseH-like_sf
IPR005828 MFS_sugar_transport-like
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR016024 ARM-type_fold
IPR000225 Armadillo
IPR011333 SKP1/BTB/POZ_sf
IPR011989 ARM-like
IPR000210 BTB/POZ_dom
IPR037120 Haem_peroxidase_sf_animal
IPR019791 Haem_peroxidase_animal
IPR010255 Haem_peroxidase_sf
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR012337 RNaseH-like_sf
IPR005828 MFS_sugar_transport-like
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR016024 ARM-type_fold
IPR000225 Armadillo
IPR011333 SKP1/BTB/POZ_sf
IPR011989 ARM-like
IPR000210 BTB/POZ_dom
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JQT0
A0A3S2NMV9
A0A0N0PED8
A0A3S2L850
A0A194Q3N0
A0A2H1W9U4
+ More
A0A3S2NLC3 A0A2H1WAX3 A0A194Q7Y0 A0A2A4IUC0 A0A194RBC8 A0A2A4J7D3 A0A194Q9H2 A0A194QDV6 A0A2W1BYE5 I4DJ10 A0A194RAP6 A0A212ETD1 A0A2W1BT67 A0A2A4IUP3 A0A2W1BQ93 A0A2A4JM77 A0A2A4JMB0 H9ISL1 H9JT77 A0A2W1BXH1 A0A2W1BS42 A0A2H1WBG2 A0A2A4IUD5 A0A2H1WGR9 A0A2A4JI21 A0A088MGW5 A0A2H1W8U7 A0A2W1BT77 H9JT78 A0A2H1VUX0 H9IT78 A0A3S2M0V1 A0A1W4WMG7 E2BCE2 N6UAQ6 D6WCM3 A0A088SIJ3 A0A194PXJ3 A0A1B6MDD3 A0A3L8DTW7 A0A1B6DIG5 A0A1B6CUT2 A0A026W3J3 A0A1B6F7C3 E2B078 A0A3S2M8S7 A0A154PJR6 A0A0L7R2K3 A0A1Y9HDW6 A0A232F2X4 A0A1Y9IWB3 A0A194RNM9 A0A182Q218 Q7QDZ3 A0A182KV49 A0A182WXU8 A0A182VL61 A0A182TM70 A0A182I5W9 K7IYZ3 A0A182PRL2 A0A310SND6 A0A182MPT8 A0A067RJX3 A0A1Y9G8K4 A0A1Y9H2I7 A0A088AE09 A0A194PVX4 A0A0P4VM15 A0A2J7RAM0 A0A182KBY5 B0WM28 B0XE84 A0A084V9X6 A0A182RQS6 W5JNS3 A0A084WCK1 A0A182IZD9 B4M5J8 T1I7W3 A0A2A3EU99 A0A0L0CRA6 A0A182Y6T5 W5JF37 A0A1Y9HES0 A0A182K431 A0A182FTE8 A0A1A9W584
A0A3S2NLC3 A0A2H1WAX3 A0A194Q7Y0 A0A2A4IUC0 A0A194RBC8 A0A2A4J7D3 A0A194Q9H2 A0A194QDV6 A0A2W1BYE5 I4DJ10 A0A194RAP6 A0A212ETD1 A0A2W1BT67 A0A2A4IUP3 A0A2W1BQ93 A0A2A4JM77 A0A2A4JMB0 H9ISL1 H9JT77 A0A2W1BXH1 A0A2W1BS42 A0A2H1WBG2 A0A2A4IUD5 A0A2H1WGR9 A0A2A4JI21 A0A088MGW5 A0A2H1W8U7 A0A2W1BT77 H9JT78 A0A2H1VUX0 H9IT78 A0A3S2M0V1 A0A1W4WMG7 E2BCE2 N6UAQ6 D6WCM3 A0A088SIJ3 A0A194PXJ3 A0A1B6MDD3 A0A3L8DTW7 A0A1B6DIG5 A0A1B6CUT2 A0A026W3J3 A0A1B6F7C3 E2B078 A0A3S2M8S7 A0A154PJR6 A0A0L7R2K3 A0A1Y9HDW6 A0A232F2X4 A0A1Y9IWB3 A0A194RNM9 A0A182Q218 Q7QDZ3 A0A182KV49 A0A182WXU8 A0A182VL61 A0A182TM70 A0A182I5W9 K7IYZ3 A0A182PRL2 A0A310SND6 A0A182MPT8 A0A067RJX3 A0A1Y9G8K4 A0A1Y9H2I7 A0A088AE09 A0A194PVX4 A0A0P4VM15 A0A2J7RAM0 A0A182KBY5 B0WM28 B0XE84 A0A084V9X6 A0A182RQS6 W5JNS3 A0A084WCK1 A0A182IZD9 B4M5J8 T1I7W3 A0A2A3EU99 A0A0L0CRA6 A0A182Y6T5 W5JF37 A0A1Y9HES0 A0A182K431 A0A182FTE8 A0A1A9W584
PDB
3R5Q
E-value=3.92738e-10,
Score=154
Ontologies
GO
PANTHER
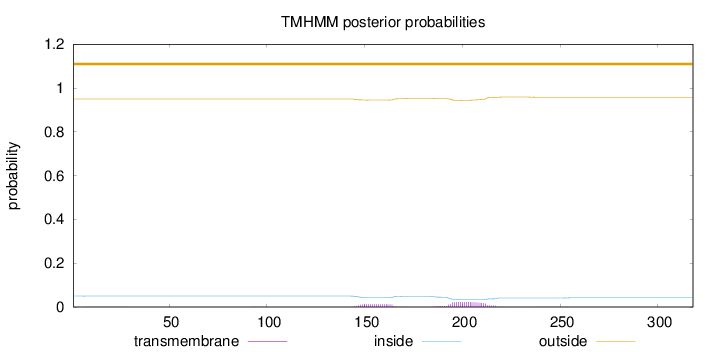
Topology
Length:
318
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.696089999999999
Exp number, first 60 AAs:
0.0086
Total prob of N-in:
0.05019
outside
1 - 318
Population Genetic Test Statistics
Pi
138.681091
Theta
162.622319
Tajima's D
-1.074726
CLR
1.795182
CSRT
0.125193740312984
Interpretation
Uncertain
