Gene
KWMTBOMO06311
Pre Gene Modal
BGIBMGA014348
Annotation
ribonuclease_E_[Moritella_viscosa]
Location in the cell
Mitochondrial Reliability : 1.207 Nuclear Reliability : 1.606
Sequence
CDS
ATGAATTGTAAAGTTGCTCTATTCCTGATAGTGGCTATTGTAGCCGTCCAGGCTCTGCCTTGCCAGGAATCTAGACCGAGACGCTGCGGGTACAGGTATGGCTGCGGAGGCGGTCGAGGCAGGGGCTCTGGTGTGAGACGTCTCGATTCAGGAGCAGATGACAACCAAGGGCGATCATGTCTCGTCGGTGGAACAGCAGCTTCGATGAGCAAAGTTCTAGCGCTAGACAAAGCAGTAGCTCGTACCAGAGCCAGTCATACAACAAAGATTCTTCAAGTACCAATGAAAGCAGTGATGGAGGCTCTGGTTCGGGCAGAACCGGTTCAGCAGGAGAAAATGGTGAGAATTCCGACGACAGCAGTGGTGCGACAAAAGGAAATAGCAGTAAATCTTCCAGCAGTTCTCAAGGTCAAAGCGCAAGTAGTAGCAGCAGTGATGAAAAATCATCTCAGAGCAGCAGTAATAGTAGCAACAACAGTAAATCAAGCAGCCAATCTTCCAGTAGCCAAAACAGTTCTGGTTCTAAGGGCTCGGGATCAGAAGAAAGCAGTAATGGAGGCTCTGGTTCGGGAAGAACCGGTTCAGCGGGAGGAACTGATGAGGATTCCGACGACAGCAGTGGTGCGACAAAAGGAAATAGCAGTAAGTCTTCCAGCAGTTCTCAAGGTCAAAGCGCAAGCAGTAGCAGCAGTGACGAAAACTCATCTCAGAGCAGCAGTAATAGTAGCAACAACAGTAAATCAAGCAGCCAATCTTCCAGTGGCCAAAACAGTTCTGGTTCTAAGGGCTCGGGATCCGAAGAAAGCAGTAATGGAGGCTCTGGTTCGGGAAGAAACGGTTCAGTGGGAGGAACTGATGAGGATTCCGACGACAGCAGTGGTGCGACAAAAGGAAATAGCAGTAAATCTTCCAGCAGCTCTCAAGGTCAAAGCGCAAGCAGTAGCAGCAGTGATGAAAAATCATCTCAGAGCAGCAGTAATAGTAGCAACAACAGTAAATCAAGCAGCCAATCTTCCAGTGGCCAAAACAGTTCTGGTTCTAAGGGCTCGGGATCAGAAGAAAGCAGTAATGGAGGCTCTGGTTCGGGAAGAAACGGTTCAGCGGGAGGAACTGATGAGGATTCCGACGACAGCAGTGGTGCGACAAAAGGAAATAGCAGTAAGTCTTCCAGCAGTTCTCAAGGTCAAAGCGCAAGCAGTAGCAGCAGTGATGAAAAATCATCTCAGAGCAGCAGTAATAGTAGCAACAACAGTAAATCAAGCAGCCAATCTTCCAGTGGCCAAAACAGTTCTGGTTCTAAGGGCTCGGGATCAGAAGAAAGCAGTAATGGAGGCTCTGGTTCGGGAAGAAACGGTTCAGCGGGAGGAACTGATGAGGATTCCGACGACAGCAGTGGTGCGACAAAAGGAAATAGCAGTAAGTCTTCCAGCAGTTCTCAAGGTCAAAGCGCAAGCAGTAGCAGCAGTGACGAAAAATCATCTCAGAGCAGCAGTAATAGTAGCAACAACAGTAAATCAAGCAGCCAATCTTCCAGTGGCCAAAACAGTTCTGGTTCTAAGGGCTCGGGATCAGAAGAAAGCAGTAATGGAGGCTCTGGTTCGGGAAGAACCGGTTCAGCGGGAGAAACTGATGAGGATTCCGACGACAGCAGTGGTGCGACAAAAGGAAATAGCAGTAAGTCTTCCAGCAGTTCTCAAGGTAAAAGTGCAAGCAGTAGCAGCAGTGACGAAAAATCATCTCAGAGTAGCAGTAATAGTAGTAACAACAGTAAATCAAGCAGTCAATCTTCGAGTAGCAACAATAGTTCTGGTTCTAAGGGCTCGGGATCAGAAGAAAGCAGTAATGGAGGCTCTGGTTCGGGAAGAACCGGTTCAGCGGGAGGAAGTGATGAGGATTCCGACGACAGCAGTGGTGCGACAAAAGGAAATAGCAGTAAGTCTTCCAGCAGCTCTCAAGGTCAAAGCGCAAGCAGTAGCAGCAGTGACGAAAAATCATCACAGAGCAACAGTAATAGTAGCAATAACAGTAAATCAAGTAGCCAATCTTCGAGTAGCAACAACAGTTCTGGTTCTAAGGGCTCGGGATCAGAAGAAAGCAGTAATGGAGGCTCTGGTTCGGGAAGAACCGGTTCAGCGGGAGGAACTGATGAGGATTCCGATGACAGCAGTGGTGCGACAAAAGGAAATAGCAGTAAGTCTTCCAGCAGTTCTCAAGGTAAAAGCGCAAGCAGTAGCAGCAGTGACGAAAAATCATCTCAGAGCAGCAGTAATAGTAGTAATAACAGTAAATCAAGCAGCCAATCGTCCAGTAGCAAGAACAGTTCTGGTTCTAAGGGCTCGGGATCAGAAGAAAGCAGTAATGGAGGCTCTGGTTCGGGAAGAACCGGTTCAGCGGGAGGAACTGATGAGGATTCCGACGACAGCAGTGGTGCGACAAAAGGAAATAGCAGTAAGTCTTCCAGCAGTTCTCAAGGTAAAAGCGCAAGCAGTAGCAGCAGTGACGAAAAATCATCTCAGAGTAACAGTAATAGTAGTAACAACAGTAAATCAAGCAGTCAATCTTCGAGTAGTAAGAACAGTTCTGGTTCTAAGGGCTCGGGATCAGAAGAAAGCAGTAATGGAGGCTCTGGTTCGGGAAGAACCGGTTCAGCGGGAGGAACTGATGAGGATTCCGACGACAGCAGTGGTGCGACAAAAGGAAATAGCAGTAAGTCTTCCAGCAGTTCTCAAGGTAAAAGCGCAAGCAGTAGCAGCAGTAACGAAAAATCATCTCAGAGTAGCAGTAATAGTAGTAACAACAGTAAATCAAGCAGTCAATCTTCGAGTAGCAAGAACAGTTCTGGTTCTAAGGGCTCTGGATCAAGTGAAAGTGGTGATAAAAAGTCCAGTTCTCGAGGAAGTTCTGGTGACAACTCAGACGATGACCAAACTGATTCAGCCAGATCAAATAGTAAGCGTTCCACAAGCTCTGATGCGTCCACTAAAAAAAGTTCGTCTAGAAAGAGCTCCAACCACCGTAGTAGCAGAAGTCAGCAAGCTCATAGTAGCAGCAGTAAACAAGCCCAAAGCAGCAGTAGTCAACAAGCCCAAAATAGCAGAAGTCAGCAAGCTCATAGTAGCAGAAGTCAGCAAGCTCATAGTAGCAGCAGTAAACAAGCCCAAAGCAGCAGTAGTAAACAAGCCCAAAGTAGCAGCAGTAAACAAGCCCAAAGCAGCAGTAGTAAACAAGCCCAAAGTAGCAGTAGTCAACAAGCCCAAAGTAGCAGAAGTCAGCAAGCTCATAGTAGCAGAAGTCAGCAAGCTCATAGTAGCAGCAGTAAACAAGCCCAAAGCAGCAGTAGTAAACAAGCCCAAAGTAGCAGCAGTAAACAAGCCCAAAGCAGCAGTAGTAAACAAGCCCAAAGCAGCAGTAGTCAACAAGCCCAAAGTAGCAGAAGTCAGCAAGCTCATAGTAGCAGAAGTGAGACAGATAGTAAGAGCAGTAATAGTGGTGGCCATTCTAATCATAGCAGTAGAACTGAACAAAAGAGCAGCGCAAAAGCAATTTCAAGCAGTGAACAGTCACAGAATTTCTCAAGCAGTTCACAGAAATCTGCCGAGGCCGCTGATGGATCTGAAAGCACCCAGTCAGCAAGTGAATACTCGAGCAGCCAATCAAAGGCCAGTAGCAGTTTTAGCGCTTCGAGTGCCAGTGAAAGTTCTTCATTAA
Protein
MNCKVALFLIVAIVAVQALPCQESRPRRCGYRYGCGGGRGRGSGVRRLDSGADDNQGRSCLVGGTAASMSKVLALDKAVARTRASHTTKILQVPMKAVMEALVRAEPVQQEKMVRIPTTAVVRQKEIAVNLPAVLKVKAQVVAAVMKNHLRAAVIVATTVNQAANLPVAKTVLVLRARDQKKAVMEALVREEPVQREELMRIPTTAVVRQKEIAVSLPAVLKVKAQAVAAVTKTHLRAAVIVATTVNQAANLPVAKTVLVLRARDPKKAVMEALVREETVQWEELMRIPTTAVVRQKEIAVNLPAALKVKAQAVAAVMKNHLRAAVIVATTVNQAANLPVAKTVLVLRARDQKKAVMEALVREETVQREELMRIPTTAVVRQKEIAVSLPAVLKVKAQAVAAVMKNHLRAAVIVATTVNQAANLPVAKTVLVLRARDQKKAVMEALVREETVQREELMRIPTTAVVRQKEIAVSLPAVLKVKAQAVAAVTKNHLRAAVIVATTVNQAANLPVAKTVLVLRARDQKKAVMEALVREEPVQREKLMRIPTTAVVRQKEIAVSLPAVLKVKVQAVAAVTKNHLRVAVIVVTTVNQAVNLRVATIVLVLRARDQKKAVMEALVREEPVQREEVMRIPTTAVVRQKEIAVSLPAALKVKAQAVAAVTKNHHRATVIVAITVNQVANLRVATTVLVLRARDQKKAVMEALVREEPVQREELMRIPMTAVVRQKEIAVSLPAVLKVKAQAVAAVTKNHLRAAVIVVITVNQAANRPVARTVLVLRARDQKKAVMEALVREEPVQREELMRIPTTAVVRQKEIAVSLPAVLKVKAQAVAAVTKNHLRVTVIVVTTVNQAVNLRVVRTVLVLRARDQKKAVMEALVREEPVQREELMRIPTTAVVRQKEIAVSLPAVLKVKAQAVAAVTKNHLRVAVIVVTTVNQAVNLRVARTVLVLRALDQVKVVIKSPVLEEVLVTTQTMTKLIQPDQIVSVPQALMRPLKKVRLERAPTTVVAEVSKLIVAAVNKPKAAVVNKPKIAEVSKLIVAEVSKLIVAAVNKPKAAVVNKPKVAAVNKPKAAVVNKPKVAVVNKPKVAEVSKLIVAEVSKLIVAAVNKPKAAVVNKPKVAAVNKPKAAVVNKPKAAVVNKPKVAEVSKLIVAEVRQIVRAVIVVAILIIAVELNKRAAQKQFQAVNSHRISQAVHRNLPRPLMDLKAPSQQVNTRAANQRPVAVLALRVPVKVLH
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
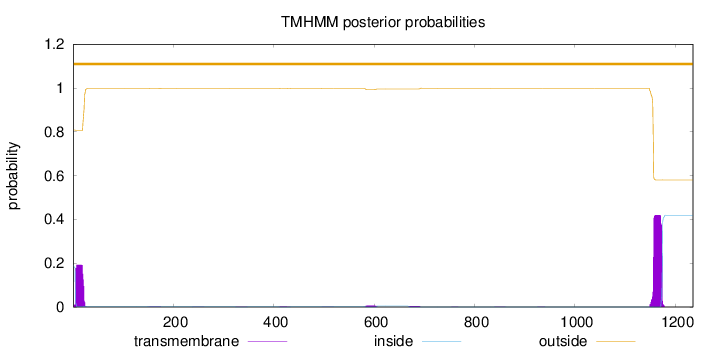
Topology
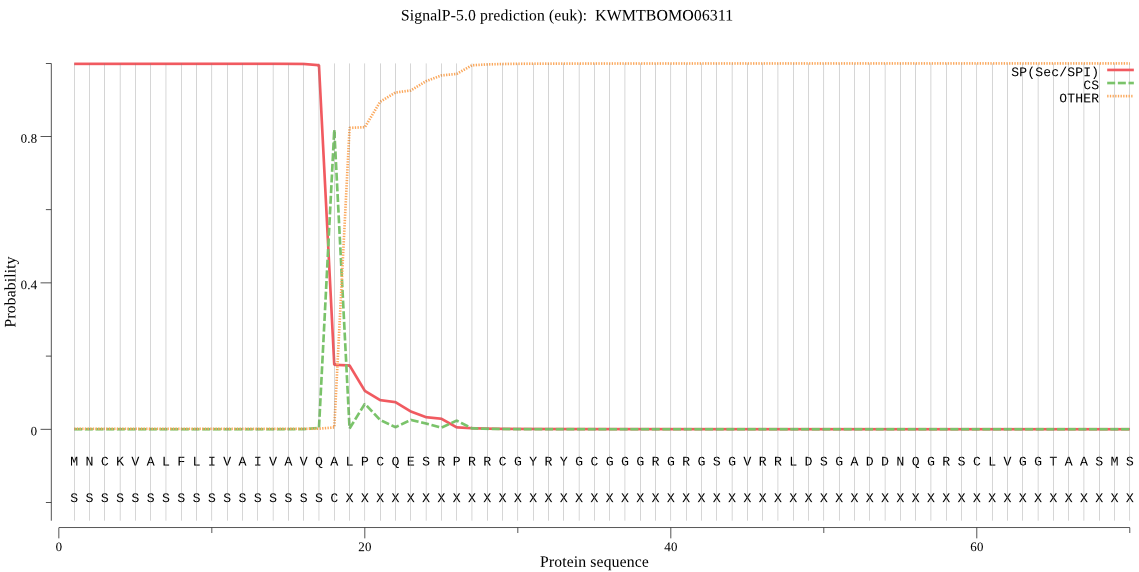
SignalP
Position: 1 - 18,
Likelihood: 0.998526
Length:
1235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.47584
Exp number, first 60 AAs:
3.31194
Total prob of N-in:
0.19166
outside
1 - 1235
Population Genetic Test Statistics
Pi
272.242334
Theta
191.088995
Tajima's D
0.973303
CLR
0.116604
CSRT
0.653017349132543
Interpretation
Uncertain
