Gene
KWMTBOMO06297
Pre Gene Modal
BGIBMGA001609
Annotation
UDP-N-acetylhexosamine_pyrophosphorylase-like_protein_1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 4.079
Sequence
CDS
ATGTATGAAACATTATTACGAAACCTAAAAGATCATGGTCAAGAACACTTAATTAAATACTGGTCGGTATTAAGTGAGGAACAGCGGAAACAGCTGTCGGACGAAATTCTAAAACTTGACTTGACGGAAGTGCATGCAACATTTTCGCGGGCAATTGAATCTACGAAAAAAATATTAGAAAAATTAGACGACGACTTGAAACCAATACCGGATAGTCACTATGAGTCTGTACCGAACTTGACGCCCGATAAGATTGAAGAATACGAAAATATCGGATTTAAAGAAATATGCAATGGGAAAGTTGGTGTGCTCTTGTTAGCTGGAGGTCAGGCGACGCGGCTCGGTTTCGGCCACCCTAAGGGAATGTACGATGTGGGATTGCCATCAAGAAAAACCTTGTTCCAGATACAAGCCGAGAGGATCCTGCGAGTTCAGCAGATGGCTGCCGAAAAATATGGAAACGAAGGTAAAATAACATGGTATATTATGACTTCTGAACACACGAAAGCCCCTACTGCAAATTATTTCAAAAGTCACTCATACTTTGGTTTGAATGAAAACGATGTGGTTTTCTTTGAGCAAGGCACATTACCTTGCTTTGATTTTGAAGGGAAAATATTCTTGGATGAGAAATATCATTTGTCTGCTGCCCCTGATGGTAATGGAGGTCTGTATCGAGCATTAAAAACCCAGGGAATATTGGACGATATCTCAGTAAGAGGAATCCAACATCTTCATGCTCATTCAGTTGACAACATACTTATAAAGGTTGCAGATCCGGTATTTATTGGTTACTGTAAATCTAAAAATGCTGATTGTGCTGCAAAAGTCGTTCAGAAATCTAGTCCCAGTGAGCCTGTTGGTGTCGTCTGCAGAGTGAATGGACACTACAAGGTAGTGGAGTACTCCGAACTCACAGACGAGGCATCTGAAAGACGTAATCCTGATGGTCGTCTCACATTCTCAGCAGGAAACATCTGTAATCATTATTTTTCAGCTGATTTCTTAAGAAAAATATCAAACTTTGAGACAAAGCTAAAATTGCATATAGCCAAAAAGAAAATACCGTATATAGATGAGAATGGTGTGCGGCAGAAACCGAATGAACCAAACGGTATTAAGATGGAGAAATTTATTTTTGATGTTTTTGAGTTTGCTGAGAATTTCATTTGCTTAGAGGTGGCTCGAGACACTGAGTTTTCAGCCTTAAAGAATGCTGACACAGCCAAAAAAGACTGTCCTTCGACTGCCCGAGAAGATTTGCTTCAGCTCCATAAAAAATACATTAGACAAGCTGGTGGAGAGGTGGCAGACGATGCTGATATAGAAATCTCTCCGTTATTGTCCTACGGGGGCGAGAATTTGGACAGCATTGTTAACGGTAAAGTGTTTACCGCTGGTCCATTCCATTTGAAGAGTCCACAGGAATTATCAAGTAATGGTGTTAATGGTAATCATTGA
Protein
MYETLLRNLKDHGQEHLIKYWSVLSEEQRKQLSDEILKLDLTEVHATFSRAIESTKKILEKLDDDLKPIPDSHYESVPNLTPDKIEEYENIGFKEICNGKVGVLLLAGGQATRLGFGHPKGMYDVGLPSRKTLFQIQAERILRVQQMAAEKYGNEGKITWYIMTSEHTKAPTANYFKSHSYFGLNENDVVFFEQGTLPCFDFEGKIFLDEKYHLSAAPDGNGGLYRALKTQGILDDISVRGIQHLHAHSVDNILIKVADPVFIGYCKSKNADCAAKVVQKSSPSEPVGVVCRVNGHYKVVEYSELTDEASERRNPDGRLTFSAGNICNHYFSADFLRKISNFETKLKLHIAKKKIPYIDENGVRQKPNEPNGIKMEKFIFDVFEFAENFICLEVARDTEFSALKNADTAKKDCPSTAREDLLQLHKKYIRQAGGEVADDADIEISPLLSYGGENLDSIVNGKVFTAGPFHLKSPQELSSNGVNGNH
Summary
Similarity
Belongs to the SecY/SEC61-alpha family.
Uniprot
H9IWH7
A0A089PRC4
A0A0H4K988
A0A2A4IW10
A0A0N1IJZ7
A0A194QAE1
+ More
A0A2W1BGK5 C0JP36 A0A2H1VI03 A0A0L7L7Z6 A0A212F0J1 A0A336MRB9 A0A1J1J4J2 A0A336K9K8 A0A1Q3FI02 B0W7S4 T1P8W1 A0A1L8E3R0 D3TMF2 Q6GW02 A0A1A9ZFD2 A0A1S4EZH5 A0A1B0BN42 A0A182GBM6 A0A1A9XQQ5 A0A0L0C2R3 A0A0M4EBZ9 B3MVM1 B4JQ73 A0A0P8ZI38 A0A0A1XIJ4 A0A1S6J0Z1 A0A3G1NF53 B4P0D5 A0A0A1WRE5 A0A0R1DP57 A0A1A9VF40 A0A0K8TDM2 A0A2K8FTL8 A0A1A9W5I7 B4M9K1 A0A146LTB7 B4N7M5 B3N5R9 A0A0A9ZAE6 A0A1W4V246 A0A0Q9WSQ0 A0A1W4VEE5 A0A2Z4BZH2 A0A1I8PSF4 A0A0Q5WJW6 A0A1I8PSE6 A0A0J9QWZ9 T1RR64 B4I1W1 A0A0J9QWR8 A0A034VRQ3 A0A0Q9XGY8 B4KKX7 A0A0K8VFV9 Q9Y0Z0 Q8IGT8 B4GKK5 Q8IPJ6 A0A034VQ51 W8CA07 W8BGG5 A0A0K8VVU0 A0A0R3NW48 Q29PE0 U5EJU3 S4TMY9 A0A3B0K9M7 A0A2P8XQZ0 A0A023F4T7 A0A2J7PZX6 A0A2R7X5X4 A0A0V0GAR1 A0A1Y1KQ16 A0A1W7R9C3 A0A1B6MLW9 A0A023ES67 A0A067R5N0 T1IG81 A0A1B6GC03 A0A0N8ER49 A0A026WN36 A0A1B6IQT1 A0A195D9D2 A0A0N0BJE1 A0A195C6V7 A0A195ERE2 J9JW65 T1RRG3 A0A2S2PAL6 F4X1T0 A0A2S2QDB6 A0A2H8TXM0
A0A2W1BGK5 C0JP36 A0A2H1VI03 A0A0L7L7Z6 A0A212F0J1 A0A336MRB9 A0A1J1J4J2 A0A336K9K8 A0A1Q3FI02 B0W7S4 T1P8W1 A0A1L8E3R0 D3TMF2 Q6GW02 A0A1A9ZFD2 A0A1S4EZH5 A0A1B0BN42 A0A182GBM6 A0A1A9XQQ5 A0A0L0C2R3 A0A0M4EBZ9 B3MVM1 B4JQ73 A0A0P8ZI38 A0A0A1XIJ4 A0A1S6J0Z1 A0A3G1NF53 B4P0D5 A0A0A1WRE5 A0A0R1DP57 A0A1A9VF40 A0A0K8TDM2 A0A2K8FTL8 A0A1A9W5I7 B4M9K1 A0A146LTB7 B4N7M5 B3N5R9 A0A0A9ZAE6 A0A1W4V246 A0A0Q9WSQ0 A0A1W4VEE5 A0A2Z4BZH2 A0A1I8PSF4 A0A0Q5WJW6 A0A1I8PSE6 A0A0J9QWZ9 T1RR64 B4I1W1 A0A0J9QWR8 A0A034VRQ3 A0A0Q9XGY8 B4KKX7 A0A0K8VFV9 Q9Y0Z0 Q8IGT8 B4GKK5 Q8IPJ6 A0A034VQ51 W8CA07 W8BGG5 A0A0K8VVU0 A0A0R3NW48 Q29PE0 U5EJU3 S4TMY9 A0A3B0K9M7 A0A2P8XQZ0 A0A023F4T7 A0A2J7PZX6 A0A2R7X5X4 A0A0V0GAR1 A0A1Y1KQ16 A0A1W7R9C3 A0A1B6MLW9 A0A023ES67 A0A067R5N0 T1IG81 A0A1B6GC03 A0A0N8ER49 A0A026WN36 A0A1B6IQT1 A0A195D9D2 A0A0N0BJE1 A0A195C6V7 A0A195ERE2 J9JW65 T1RRG3 A0A2S2PAL6 F4X1T0 A0A2S2QDB6 A0A2H8TXM0
Pubmed
19121390
26354079
28756777
26227816
22118469
25315136
+ More
20353571 17510324 26483478 26108605 17994087 18057021 25830018 27414796 17550304 26823975 25401762 22936249 23977188 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085 29403074 25474469 28004739 24945155 24845553 24508170 30249741 21719571
20353571 17510324 26483478 26108605 17994087 18057021 25830018 27414796 17550304 26823975 25401762 22936249 23977188 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085 29403074 25474469 28004739 24945155 24845553 24508170 30249741 21719571
EMBL
BABH01021347
KM078925
AIQ85099.1
KP705074
AKO90063.1
NWSH01005743
+ More
PCG63995.1 KQ459736 KPJ20218.1 KQ459463 KPJ00381.1 KZ150077 PZC73938.1 FJ380203 ACN29686.1 ODYU01002647 SOQ40426.1 JTDY01002443 KOB71416.1 AGBW02011074 OWR47250.1 UFQS01001884 UFQT01001884 SSX12648.1 SSX32091.1 CVRI01000070 CRL07311.1 UFQS01000123 UFQT01000123 SSX00065.1 SSX20445.1 GFDL01007844 JAV27201.1 DS231855 EDS38218.1 KA644575 AFP59204.1 GFDF01000825 JAV13259.1 CCAG010016184 EZ422604 ADD18880.1 AY631402 AY632694 CH477222 AAT48092.1 EAT47260.1 JXJN01017195 JXUM01053044 KQ561760 KXJ77610.1 JRES01000973 KNC26643.1 CP012523 ALC40214.1 CH902624 EDV33286.1 CH916372 EDV99053.1 KPU74384.1 KPU74385.1 GBXI01003506 JAD10786.1 KU764442 AQS60688.1 MF964941 AUX15121.1 CM000157 EDW87893.1 GBXI01012900 JAD01392.1 KRJ97474.1 GBRD01002222 JAG63599.1 KU556836 KY618822 AQT25651.1 AQW43008.1 CH940654 EDW57877.2 GDHC01007645 JAQ10984.1 CH964214 EDW80364.1 CH954177 EDV59078.1 GBHO01043421 GBHO01043420 GBHO01043418 GBHO01002733 GBHO01000227 GBHO01000225 JAG00183.1 JAG00184.1 JAG00186.1 JAG40871.1 JAG43377.1 JAG43379.1 KRF99269.1 MH119582 AWU68200.1 KQS70619.1 CM002910 KMY88523.1 JX484802 AGN56418.1 CH480820 EDW54518.1 KMY88522.1 GAKP01014754 JAC44198.1 CH933807 KRG02856.1 EDW11707.2 GDHF01014577 JAI37737.1 AF145686 AE014134 AAD38661.1 AAF52387.1 BT001608 AAN71363.1 CH479184 EDW37171.1 AAN10586.1 AHN54188.1 GAKP01014751 GAKP01014750 JAC44202.1 GAMC01006191 JAC00365.1 GAMC01006190 JAC00366.1 GDHF01009317 JAI42997.1 CH379058 KRT03421.1 EAL34353.1 GANO01002095 JAB57776.1 JX434756 AGE89783.1 OUUW01000064 SPP90063.1 PYGN01001499 PSN34428.1 GBBI01002689 JAC16023.1 NEVH01020330 PNF21887.1 KK857445 PTY27176.1 GECL01001783 JAP04341.1 GEZM01076968 JAV63502.1 GFAH01000664 JAV47725.1 GEBQ01003041 JAT36936.1 GAPW01001510 JAC12088.1 KK852869 KDR14652.1 ACPB03002356 GECZ01009804 JAS59965.1 GDIQ01002076 JAN92661.1 KK107152 QOIP01000007 EZA57056.1 RLU20461.1 GECU01018446 GECU01002970 JAS89260.1 JAT04737.1 KQ981153 KYN09009.1 KQ435716 KOX79054.1 KQ978251 KYM95903.1 KQ981993 KYN30825.1 ABLF02036121 JX484803 AGN56419.1 GGMR01013609 MBY26228.1 GL888551 EGI59587.1 GGMS01006468 MBY75671.1 GFXV01006467 MBW18272.1
PCG63995.1 KQ459736 KPJ20218.1 KQ459463 KPJ00381.1 KZ150077 PZC73938.1 FJ380203 ACN29686.1 ODYU01002647 SOQ40426.1 JTDY01002443 KOB71416.1 AGBW02011074 OWR47250.1 UFQS01001884 UFQT01001884 SSX12648.1 SSX32091.1 CVRI01000070 CRL07311.1 UFQS01000123 UFQT01000123 SSX00065.1 SSX20445.1 GFDL01007844 JAV27201.1 DS231855 EDS38218.1 KA644575 AFP59204.1 GFDF01000825 JAV13259.1 CCAG010016184 EZ422604 ADD18880.1 AY631402 AY632694 CH477222 AAT48092.1 EAT47260.1 JXJN01017195 JXUM01053044 KQ561760 KXJ77610.1 JRES01000973 KNC26643.1 CP012523 ALC40214.1 CH902624 EDV33286.1 CH916372 EDV99053.1 KPU74384.1 KPU74385.1 GBXI01003506 JAD10786.1 KU764442 AQS60688.1 MF964941 AUX15121.1 CM000157 EDW87893.1 GBXI01012900 JAD01392.1 KRJ97474.1 GBRD01002222 JAG63599.1 KU556836 KY618822 AQT25651.1 AQW43008.1 CH940654 EDW57877.2 GDHC01007645 JAQ10984.1 CH964214 EDW80364.1 CH954177 EDV59078.1 GBHO01043421 GBHO01043420 GBHO01043418 GBHO01002733 GBHO01000227 GBHO01000225 JAG00183.1 JAG00184.1 JAG00186.1 JAG40871.1 JAG43377.1 JAG43379.1 KRF99269.1 MH119582 AWU68200.1 KQS70619.1 CM002910 KMY88523.1 JX484802 AGN56418.1 CH480820 EDW54518.1 KMY88522.1 GAKP01014754 JAC44198.1 CH933807 KRG02856.1 EDW11707.2 GDHF01014577 JAI37737.1 AF145686 AE014134 AAD38661.1 AAF52387.1 BT001608 AAN71363.1 CH479184 EDW37171.1 AAN10586.1 AHN54188.1 GAKP01014751 GAKP01014750 JAC44202.1 GAMC01006191 JAC00365.1 GAMC01006190 JAC00366.1 GDHF01009317 JAI42997.1 CH379058 KRT03421.1 EAL34353.1 GANO01002095 JAB57776.1 JX434756 AGE89783.1 OUUW01000064 SPP90063.1 PYGN01001499 PSN34428.1 GBBI01002689 JAC16023.1 NEVH01020330 PNF21887.1 KK857445 PTY27176.1 GECL01001783 JAP04341.1 GEZM01076968 JAV63502.1 GFAH01000664 JAV47725.1 GEBQ01003041 JAT36936.1 GAPW01001510 JAC12088.1 KK852869 KDR14652.1 ACPB03002356 GECZ01009804 JAS59965.1 GDIQ01002076 JAN92661.1 KK107152 QOIP01000007 EZA57056.1 RLU20461.1 GECU01018446 GECU01002970 JAS89260.1 JAT04737.1 KQ981153 KYN09009.1 KQ435716 KOX79054.1 KQ978251 KYM95903.1 KQ981993 KYN30825.1 ABLF02036121 JX484803 AGN56419.1 GGMR01013609 MBY26228.1 GL888551 EGI59587.1 GGMS01006468 MBY75671.1 GFXV01006467 MBW18272.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000183832 UP000002320 UP000095301 UP000092444 UP000008820 UP000092445 UP000092460 UP000069940 UP000249989 UP000092443 UP000037069 UP000092553 UP000007801 UP000001070 UP000002282 UP000078200 UP000091820 UP000008792 UP000007798 UP000008711 UP000192221 UP000095300 UP000001292 UP000009192 UP000000803 UP000008744 UP000001819 UP000268350 UP000245037 UP000235965 UP000027135 UP000015103 UP000053097 UP000279307 UP000078492 UP000053105 UP000078542 UP000078541 UP000007819 UP000007755
UP000183832 UP000002320 UP000095301 UP000092444 UP000008820 UP000092445 UP000092460 UP000069940 UP000249989 UP000092443 UP000037069 UP000092553 UP000007801 UP000001070 UP000002282 UP000078200 UP000091820 UP000008792 UP000007798 UP000008711 UP000192221 UP000095300 UP000001292 UP000009192 UP000000803 UP000008744 UP000001819 UP000268350 UP000245037 UP000235965 UP000027135 UP000015103 UP000053097 UP000279307 UP000078492 UP000053105 UP000078542 UP000078541 UP000007819 UP000007755
Interpro
Gene 3D
ProteinModelPortal
H9IWH7
A0A089PRC4
A0A0H4K988
A0A2A4IW10
A0A0N1IJZ7
A0A194QAE1
+ More
A0A2W1BGK5 C0JP36 A0A2H1VI03 A0A0L7L7Z6 A0A212F0J1 A0A336MRB9 A0A1J1J4J2 A0A336K9K8 A0A1Q3FI02 B0W7S4 T1P8W1 A0A1L8E3R0 D3TMF2 Q6GW02 A0A1A9ZFD2 A0A1S4EZH5 A0A1B0BN42 A0A182GBM6 A0A1A9XQQ5 A0A0L0C2R3 A0A0M4EBZ9 B3MVM1 B4JQ73 A0A0P8ZI38 A0A0A1XIJ4 A0A1S6J0Z1 A0A3G1NF53 B4P0D5 A0A0A1WRE5 A0A0R1DP57 A0A1A9VF40 A0A0K8TDM2 A0A2K8FTL8 A0A1A9W5I7 B4M9K1 A0A146LTB7 B4N7M5 B3N5R9 A0A0A9ZAE6 A0A1W4V246 A0A0Q9WSQ0 A0A1W4VEE5 A0A2Z4BZH2 A0A1I8PSF4 A0A0Q5WJW6 A0A1I8PSE6 A0A0J9QWZ9 T1RR64 B4I1W1 A0A0J9QWR8 A0A034VRQ3 A0A0Q9XGY8 B4KKX7 A0A0K8VFV9 Q9Y0Z0 Q8IGT8 B4GKK5 Q8IPJ6 A0A034VQ51 W8CA07 W8BGG5 A0A0K8VVU0 A0A0R3NW48 Q29PE0 U5EJU3 S4TMY9 A0A3B0K9M7 A0A2P8XQZ0 A0A023F4T7 A0A2J7PZX6 A0A2R7X5X4 A0A0V0GAR1 A0A1Y1KQ16 A0A1W7R9C3 A0A1B6MLW9 A0A023ES67 A0A067R5N0 T1IG81 A0A1B6GC03 A0A0N8ER49 A0A026WN36 A0A1B6IQT1 A0A195D9D2 A0A0N0BJE1 A0A195C6V7 A0A195ERE2 J9JW65 T1RRG3 A0A2S2PAL6 F4X1T0 A0A2S2QDB6 A0A2H8TXM0
A0A2W1BGK5 C0JP36 A0A2H1VI03 A0A0L7L7Z6 A0A212F0J1 A0A336MRB9 A0A1J1J4J2 A0A336K9K8 A0A1Q3FI02 B0W7S4 T1P8W1 A0A1L8E3R0 D3TMF2 Q6GW02 A0A1A9ZFD2 A0A1S4EZH5 A0A1B0BN42 A0A182GBM6 A0A1A9XQQ5 A0A0L0C2R3 A0A0M4EBZ9 B3MVM1 B4JQ73 A0A0P8ZI38 A0A0A1XIJ4 A0A1S6J0Z1 A0A3G1NF53 B4P0D5 A0A0A1WRE5 A0A0R1DP57 A0A1A9VF40 A0A0K8TDM2 A0A2K8FTL8 A0A1A9W5I7 B4M9K1 A0A146LTB7 B4N7M5 B3N5R9 A0A0A9ZAE6 A0A1W4V246 A0A0Q9WSQ0 A0A1W4VEE5 A0A2Z4BZH2 A0A1I8PSF4 A0A0Q5WJW6 A0A1I8PSE6 A0A0J9QWZ9 T1RR64 B4I1W1 A0A0J9QWR8 A0A034VRQ3 A0A0Q9XGY8 B4KKX7 A0A0K8VFV9 Q9Y0Z0 Q8IGT8 B4GKK5 Q8IPJ6 A0A034VQ51 W8CA07 W8BGG5 A0A0K8VVU0 A0A0R3NW48 Q29PE0 U5EJU3 S4TMY9 A0A3B0K9M7 A0A2P8XQZ0 A0A023F4T7 A0A2J7PZX6 A0A2R7X5X4 A0A0V0GAR1 A0A1Y1KQ16 A0A1W7R9C3 A0A1B6MLW9 A0A023ES67 A0A067R5N0 T1IG81 A0A1B6GC03 A0A0N8ER49 A0A026WN36 A0A1B6IQT1 A0A195D9D2 A0A0N0BJE1 A0A195C6V7 A0A195ERE2 J9JW65 T1RRG3 A0A2S2PAL6 F4X1T0 A0A2S2QDB6 A0A2H8TXM0
PDB
1JV3
E-value=2.10874e-134,
Score=1228
Ontologies
PATHWAY
GO
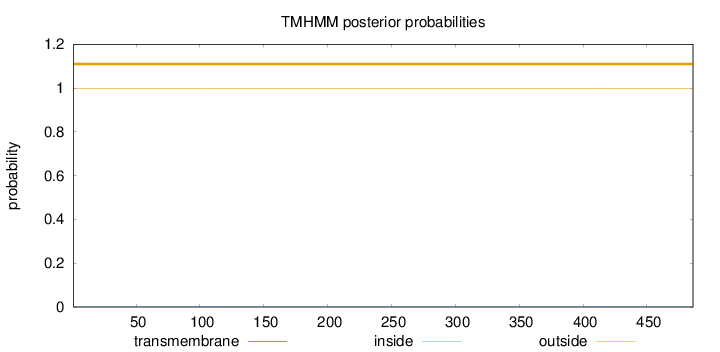
Topology
Subcellular location
Membrane
Length:
486
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00892
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00329
outside
1 - 486
Population Genetic Test Statistics
Pi
230.753763
Theta
185.27127
Tajima's D
0.892467
CLR
0
CSRT
0.634818259087046
Interpretation
Uncertain
