Gene
KWMTBOMO06296
Pre Gene Modal
BGIBMGA001610
Annotation
PREDICTED:_regulator_of_G-protein_signaling_7_[Papilio_xuthus]
Full name
MICOS complex subunit
Location in the cell
Nuclear Reliability : 2.816
Sequence
CDS
ATGGTTACGATGAATACTACGGAAAAGGCGCCGAAGCCCTGTCAGCAAGGTCCATCAAGGAATAACTCAAAGTGTGTTAAAGAAGAGAAAGACAAGGAGAAGCGAGATAATGCCATTACTAACTTGCCACCGAGCGTCCAGGCCCTTATGGCTGCAATACCTTCACCCGAGGAATCACCAAACTATTTGATTTACAAAAAGATGGAAGCTTTGATTGAGAAAATGCAAGATGAGAACACGGGAGTACCTGTTCGAACTGTGAAAAGCTTTATGACGAAAGTTCCATCGGTGTTCACCGGCGCAGAGCTGGTGTCGTGGCTGGCAAGGCATCTGCAACTTGACGAGCCCTGGGAACCACTGCACCTTGCCCATCTCCTGGCTGCCCACGGCTATTTATTTCCCATAGACGATCATTGTTTGACCGTTAAAAACGATAACACGTATTATAGATTCCAGACCCCGTATTTCTGGCCTTCGAACCAATGGGAGCCTGAGAATACTGATTACGCGGTGTACCTCTGCAAGCGGACGATGCAGAACAAGGCGCGGCTAGAGCTGGCGGACTATGAGGCCGAATCATTAGCGAAACTCCAGAAGATGTTTTCGAGAAAATGGGAATTCATATTCATGCAAGCCGAGGTTGACAAGAAACGTGATAAGTTGGAGAGGAAGGTGCTGGACAGTCAGGAGAGGGCCTTCTGGGACGTGCATCGACCGATGCCTGGTTGCGTGAACACTACAGAGTTAGATCCGAGGAAGCTTTCTCGTATAAATGCGCTTAAAGCTAAACGTCTCAAACAATGTCAGGACTTGGTGCTGCAGAACAACTTGAAGAACAATCCAATTGTGACCATAACGAACGCCCAGGAAAGCGAAGATGAGATCCAGAGGAACAACCAGGAGAACGTCATCAAAGAGAATGGTATTTTTTATTTATTGCTTAGATGGGTGGACGAGCTCACGGTCCACCTGGCATTAAGTGGTCACCGGAGCCCGTAG
Protein
MVTMNTTEKAPKPCQQGPSRNNSKCVKEEKDKEKRDNAITNLPPSVQALMAAIPSPEESPNYLIYKKMEALIEKMQDENTGVPVRTVKSFMTKVPSVFTGAELVSWLARHLQLDEPWEPLHLAHLLAAHGYLFPIDDHCLTVKNDNTYYRFQTPYFWPSNQWEPENTDYAVYLCKRTMQNKARLELADYEAESLAKLQKMFSRKWEFIFMQAEVDKKRDKLERKVLDSQERAFWDVHRPMPGCVNTTELDPRKLSRINALKAKRLKQCQDLVLQNNLKNNPIVTITNAQESEDEIQRNNQENVIKENGIFYLLLRWVDELTVHLALSGHRSP
Summary
Description
Component of the MICOS complex, a large protein complex of the mitochondrial inner membrane that plays crucial roles in the maintenance of crista junctions, inner membrane architecture, and formation of contact sites to the outer membrane.
Subunit
Component of the mitochondrial contact site and cristae organizing system (MICOS) complex.
Uniprot
H9IWH8
A0A194Q4B2
A0A2H1VHX1
E2BZR8
A0A0A9XQ90
A0A026WEE3
+ More
A0A1Y1N895 D6W688 A0A182SXS2 A0A0C9R4T5 A0A0C9RRW6 A0A0M8ZMQ9 A0A087ZT40 E2A3N4 A0A1Y1N739 A0A151JT63 N6TY94 F4W7F9 A0A182PQQ8 A0A182JVQ1 A0A182V8Y1 Q7QG26 A0A182WVD6 A0A151X8L8 A0A182I917 A0A182LAZ8 A0A182UGW6 A0A1B0CHC5 A0A0L7RDX7 A0A151I6X6 A0A2A3E8I6 K7J2P9
A0A1Y1N895 D6W688 A0A182SXS2 A0A0C9R4T5 A0A0C9RRW6 A0A0M8ZMQ9 A0A087ZT40 E2A3N4 A0A1Y1N739 A0A151JT63 N6TY94 F4W7F9 A0A182PQQ8 A0A182JVQ1 A0A182V8Y1 Q7QG26 A0A182WVD6 A0A151X8L8 A0A182I917 A0A182LAZ8 A0A182UGW6 A0A1B0CHC5 A0A0L7RDX7 A0A151I6X6 A0A2A3E8I6 K7J2P9
Pubmed
EMBL
BABH01021340
BABH01021341
KQ459463
KPJ00382.1
ODYU01002647
SOQ40430.1
+ More
GL451657 EFN78830.1 GBHO01020692 JAG22912.1 KK107249 QOIP01000013 EZA54470.1 RLU15191.1 GEZM01013540 JAV92496.1 KQ971306 EFA11094.1 GBYB01011296 GBYB01011299 JAG81063.1 JAG81066.1 GBYB01011298 GBYB01011300 JAG81065.1 JAG81067.1 KQ436240 KOX67385.1 GL436457 EFN71926.1 GEZM01013541 JAV92495.1 KQ982014 KYN30646.1 APGK01047258 KB741077 ENN74290.1 GL887844 EGI69832.1 AAAB01008844 EAA05965.5 KQ982409 KYQ56713.1 APCN01003158 AJWK01012212 AJWK01012213 KQ414612 KOC69187.1 KQ978456 KYM93884.1 KZ288349 PBC27371.1
GL451657 EFN78830.1 GBHO01020692 JAG22912.1 KK107249 QOIP01000013 EZA54470.1 RLU15191.1 GEZM01013540 JAV92496.1 KQ971306 EFA11094.1 GBYB01011296 GBYB01011299 JAG81063.1 JAG81066.1 GBYB01011298 GBYB01011300 JAG81065.1 JAG81067.1 KQ436240 KOX67385.1 GL436457 EFN71926.1 GEZM01013541 JAV92495.1 KQ982014 KYN30646.1 APGK01047258 KB741077 ENN74290.1 GL887844 EGI69832.1 AAAB01008844 EAA05965.5 KQ982409 KYQ56713.1 APCN01003158 AJWK01012212 AJWK01012213 KQ414612 KOC69187.1 KQ978456 KYM93884.1 KZ288349 PBC27371.1
Proteomes
UP000005204
UP000053268
UP000008237
UP000053097
UP000279307
UP000007266
+ More
UP000075901 UP000053105 UP000005203 UP000000311 UP000078541 UP000019118 UP000007755 UP000075885 UP000075881 UP000075903 UP000007062 UP000076407 UP000075809 UP000075840 UP000075882 UP000075902 UP000092461 UP000053825 UP000078542 UP000242457 UP000002358
UP000075901 UP000053105 UP000005203 UP000000311 UP000078541 UP000019118 UP000007755 UP000075885 UP000075881 UP000075903 UP000007062 UP000076407 UP000075809 UP000075840 UP000075882 UP000075902 UP000092461 UP000053825 UP000078542 UP000242457 UP000002358
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9IWH8
A0A194Q4B2
A0A2H1VHX1
E2BZR8
A0A0A9XQ90
A0A026WEE3
+ More
A0A1Y1N895 D6W688 A0A182SXS2 A0A0C9R4T5 A0A0C9RRW6 A0A0M8ZMQ9 A0A087ZT40 E2A3N4 A0A1Y1N739 A0A151JT63 N6TY94 F4W7F9 A0A182PQQ8 A0A182JVQ1 A0A182V8Y1 Q7QG26 A0A182WVD6 A0A151X8L8 A0A182I917 A0A182LAZ8 A0A182UGW6 A0A1B0CHC5 A0A0L7RDX7 A0A151I6X6 A0A2A3E8I6 K7J2P9
A0A1Y1N895 D6W688 A0A182SXS2 A0A0C9R4T5 A0A0C9RRW6 A0A0M8ZMQ9 A0A087ZT40 E2A3N4 A0A1Y1N739 A0A151JT63 N6TY94 F4W7F9 A0A182PQQ8 A0A182JVQ1 A0A182V8Y1 Q7QG26 A0A182WVD6 A0A151X8L8 A0A182I917 A0A182LAZ8 A0A182UGW6 A0A1B0CHC5 A0A0L7RDX7 A0A151I6X6 A0A2A3E8I6 K7J2P9
PDB
6N9G
E-value=7.33642e-81,
Score=764
Ontologies
KEGG
PANTHER
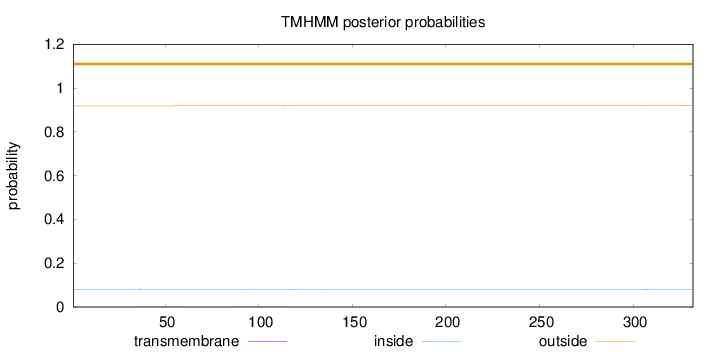
Topology
Subcellular location
Mitochondrion inner membrane
Length:
332
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02958
Exp number, first 60 AAs:
0.00478
Total prob of N-in:
0.08145
outside
1 - 332
Population Genetic Test Statistics
Pi
157.146554
Theta
151.388052
Tajima's D
0.484498
CLR
0.009746
CSRT
0.512774361281936
Interpretation
Uncertain
