Gene
KWMTBOMO06295
Pre Gene Modal
BGIBMGA001610
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_regulator_of_G-protein_signaling_7_[Amyelois_transitella]
Full name
MICOS complex subunit
Location in the cell
Cytoplasmic Reliability : 1.223 Mitochondrial Reliability : 1.809 Nuclear Reliability : 1.219
Sequence
CDS
ATGTGCTTGGTTCTAAGGGAATCGTCAAAGGAGATTCCGTCGGGTTCCACTGACCCCGCAGTGAACATCGAACAGGAGATAGAGGCGGCACGAAAGCAGCTAGGGATGCTGCGAGCGAGAATGGAACGGCGCAACATTAGGGTCAGCAAAGTCGCCGAAACGTTCGTGTCGTACTGCGAGCAGTATGCGGAGTACGACGCGTTCCTGACGCCCCCGGAGTGGCCCAACCCCTGGATCAACTACACCACCGAGCTGTGGGAGCAGGAGAAGCAATCCAAGGACCCCTTACCGTTGCGGCGGGTCCGTCGCTGGTCGTTCGGGCTGCACGAGCTGCTGCAGGATCCGGCCGGAAAGGAGCACTTCGCGAAGTTCCTCTCCAAAGAGTTCAGCGGCGAGAATCTCAAGTTCTGGCTGGCGGTGCAGGACCTGAAGGCACTGCCCATCCGCTGCGTGGCGGCGCGCGCCCGCGACATCTGGAAGGAGTTCCTCGCGGCGGACGCGCCCTCGCCCATCAATATCGACGCGGCCAGCAGGGAACTGACCCGCGTGCAGGTCGAGTCGGGCACGGCTGACCGGTACTGCTTCGATCAGGCGCAGGCTCACGTGTATCACCTAATGAAGTCCGATAGCTACTCGAGATACATCCGATCGGAAATGTACAAGGAGTACATGAATGGCTCCAAGAAGAAGACTTCAGTCAAAGGGATTCGTTCGATTGTATCGTTCACGGCGCGGAAGGAGGCGACGGCCACATAA
Protein
MCLVLRESSKEIPSGSTDPAVNIEQEIEAARKQLGMLRARMERRNIRVSKVAETFVSYCEQYAEYDAFLTPPEWPNPWINYTTELWEQEKQSKDPLPLRRVRRWSFGLHELLQDPAGKEHFAKFLSKEFSGENLKFWLAVQDLKALPIRCVAARARDIWKEFLAADAPSPINIDAASRELTRVQVESGTADRYCFDQAQAHVYHLMKSDSYSRYIRSEMYKEYMNGSKKKTSVKGIRSIVSFTARKEATAT
Summary
Description
Component of the MICOS complex, a large protein complex of the mitochondrial inner membrane that plays crucial roles in the maintenance of crista junctions, inner membrane architecture, and formation of contact sites to the outer membrane.
Subunit
Component of the mitochondrial contact site and cristae organizing system (MICOS) complex.
Uniprot
A0A2A4JI50
A0A2H1VHX1
A0A2W1BJW2
D6W688
A0A067R802
A0A2J7R1W9
+ More
A0A1B6I5F6 A0A1W4XFI9 A0A1B6MK51 A0A1B6CRF9 A0A1B6EHR9 A0A1Y1NA99 A0A1Y1N739 A0A3Q0J0K6 A0A1B6DTC2 E2BZR8 A0A087TF13 A0A151X8L8 E2A3N4 A0A023F2T9 A0A087ZT40 E9IX31 A0A026WEE3 A0A0C9RRW6 A0A158NPT6 A0A2L2YV77 A0A1Q3FQE7 A0A2R5LD80 F4W7F9 A0A1S4FAF9 J9JRI6 T1HTW9 A0A151I6X6 A0A195EM14 A0A151JT63 A0A1B0CHC5 E0VV64 A0A1B0D9B8 T1KYN9 A0A2H8TR24 A0A182NCM2 A0A195ATE8 A0A336LQ91 N6TY94 A0A182FEA3 A0A084WMH0 A0A182V8Y1 A0A182QVX1 A0A182J744 Q7QG26 A0A182WVD6 A0A232EEP4 W5JL55 B0X6J3 A0A1Q3FQC6 W5JR10 Q16SA0 Q17A55 A0A182GGD2 A0A182GMN1 A0A182H982 A0A182K6H8 A0A182NI78 A0A182M3S7 A0A182PDL6 A0A182R976 A0A182QGX7 A0A182UJX7 A0A182VLX7 A0A0P5D9S1 A0A182I4K8 A0A182X4L1 A0A182LKC2 Q7Q555 A0A182Y2H9 A0A0L7RDX7 A0A154P5X7 A0A0C9R4T5 A0A182WBN8 A0A182JVQ1 A0A182PQQ8 A0A182YJL9 A0A084WR18 A0A224XU05 A0A164MNX4 A0A182LRG3 A0A182W313 A0A182SJE8 A0A182F8F9 A0A182UGW6 B0WMV2 A0A182I917 A0A1E1XJR3 A0A182LAZ8 A0A182J8R6 V4A1D0 A0A0P5JC79 A0A034V4Z1
A0A1B6I5F6 A0A1W4XFI9 A0A1B6MK51 A0A1B6CRF9 A0A1B6EHR9 A0A1Y1NA99 A0A1Y1N739 A0A3Q0J0K6 A0A1B6DTC2 E2BZR8 A0A087TF13 A0A151X8L8 E2A3N4 A0A023F2T9 A0A087ZT40 E9IX31 A0A026WEE3 A0A0C9RRW6 A0A158NPT6 A0A2L2YV77 A0A1Q3FQE7 A0A2R5LD80 F4W7F9 A0A1S4FAF9 J9JRI6 T1HTW9 A0A151I6X6 A0A195EM14 A0A151JT63 A0A1B0CHC5 E0VV64 A0A1B0D9B8 T1KYN9 A0A2H8TR24 A0A182NCM2 A0A195ATE8 A0A336LQ91 N6TY94 A0A182FEA3 A0A084WMH0 A0A182V8Y1 A0A182QVX1 A0A182J744 Q7QG26 A0A182WVD6 A0A232EEP4 W5JL55 B0X6J3 A0A1Q3FQC6 W5JR10 Q16SA0 Q17A55 A0A182GGD2 A0A182GMN1 A0A182H982 A0A182K6H8 A0A182NI78 A0A182M3S7 A0A182PDL6 A0A182R976 A0A182QGX7 A0A182UJX7 A0A182VLX7 A0A0P5D9S1 A0A182I4K8 A0A182X4L1 A0A182LKC2 Q7Q555 A0A182Y2H9 A0A0L7RDX7 A0A154P5X7 A0A0C9R4T5 A0A182WBN8 A0A182JVQ1 A0A182PQQ8 A0A182YJL9 A0A084WR18 A0A224XU05 A0A164MNX4 A0A182LRG3 A0A182W313 A0A182SJE8 A0A182F8F9 A0A182UGW6 B0WMV2 A0A182I917 A0A1E1XJR3 A0A182LAZ8 A0A182J8R6 V4A1D0 A0A0P5JC79 A0A034V4Z1
Pubmed
EMBL
NWSH01001380
PCG71469.1
ODYU01002647
SOQ40430.1
KZ150077
PZC73934.1
+ More
KQ971306 EFA11094.1 KK852818 KDR15671.1 NEVH01008202 PNF34822.1 GECU01025548 JAS82158.1 GEBQ01003630 JAT36347.1 GEDC01021301 JAS15997.1 GECZ01032283 JAS37486.1 GEZM01013539 JAV92497.1 GEZM01013541 JAV92495.1 GEDC01024324 GEDC01018476 GEDC01018301 GEDC01008372 GEDC01006956 GEDC01000376 JAS12974.1 JAS18822.1 JAS18997.1 JAS28926.1 JAS30342.1 JAS36922.1 GL451657 EFN78830.1 KK114920 KFM63702.1 KQ982409 KYQ56713.1 GL436457 EFN71926.1 GBBI01002780 JAC15932.1 GL766616 EFZ14872.1 KK107249 QOIP01000013 EZA54470.1 RLU15191.1 GBYB01011298 GBYB01011300 JAG81065.1 JAG81067.1 ADTU01022745 ADTU01022746 IAAA01065938 LAA11929.1 GFDL01005392 JAV29653.1 GGLE01003310 MBY07436.1 GL887844 EGI69832.1 ABLF02009799 ACPB03024220 KQ978456 KYM93884.1 KQ978730 KYN28919.1 KQ982014 KYN30646.1 AJWK01012212 AJWK01012213 DS235803 EEB17270.1 AJVK01004315 CAEY01000714 GFXV01004800 MBW16605.1 KQ976745 KYM75466.1 UFQS01000111 UFQT01000111 SSW99802.1 SSX20182.1 APGK01047258 KB741077 ENN74290.1 ATLV01024452 KE525352 KFB51414.1 AXCN02001183 AAAB01008844 EAA05965.5 NNAY01005290 OXU16782.1 ADMH02001252 ETN63494.1 DS232415 EDS41476.1 GFDL01005412 JAV29633.1 ADMH02000584 ETN65743.1 CH477680 EAT37315.1 CH477340 EAT43109.1 JXUM01061490 JXUM01061491 JXUM01061492 JXUM01061493 KQ562152 KXJ76557.1 JXUM01075103 JXUM01075104 KQ562869 KXJ74964.1 JXUM01120070 KQ566355 KXJ70190.1 AXCM01002234 AXCN02000483 GDIP01175338 JAJ48064.1 APCN01002316 AAAB01008960 EAA10807.3 KQ414612 KOC69187.1 KQ434823 KZC07253.1 GBYB01011296 GBYB01011299 JAG81063.1 JAG81066.1 ATLV01025888 KE525402 KFB52662.1 GFTR01004843 JAW11583.1 LRGB01002992 KZS05223.1 AXCM01003114 DS232003 EDS31344.1 APCN01003158 GFAA01004000 JAT99434.1 KB202640 ESO88740.1 GDIQ01204199 JAK47526.1 GAKP01021363 JAC37589.1
KQ971306 EFA11094.1 KK852818 KDR15671.1 NEVH01008202 PNF34822.1 GECU01025548 JAS82158.1 GEBQ01003630 JAT36347.1 GEDC01021301 JAS15997.1 GECZ01032283 JAS37486.1 GEZM01013539 JAV92497.1 GEZM01013541 JAV92495.1 GEDC01024324 GEDC01018476 GEDC01018301 GEDC01008372 GEDC01006956 GEDC01000376 JAS12974.1 JAS18822.1 JAS18997.1 JAS28926.1 JAS30342.1 JAS36922.1 GL451657 EFN78830.1 KK114920 KFM63702.1 KQ982409 KYQ56713.1 GL436457 EFN71926.1 GBBI01002780 JAC15932.1 GL766616 EFZ14872.1 KK107249 QOIP01000013 EZA54470.1 RLU15191.1 GBYB01011298 GBYB01011300 JAG81065.1 JAG81067.1 ADTU01022745 ADTU01022746 IAAA01065938 LAA11929.1 GFDL01005392 JAV29653.1 GGLE01003310 MBY07436.1 GL887844 EGI69832.1 ABLF02009799 ACPB03024220 KQ978456 KYM93884.1 KQ978730 KYN28919.1 KQ982014 KYN30646.1 AJWK01012212 AJWK01012213 DS235803 EEB17270.1 AJVK01004315 CAEY01000714 GFXV01004800 MBW16605.1 KQ976745 KYM75466.1 UFQS01000111 UFQT01000111 SSW99802.1 SSX20182.1 APGK01047258 KB741077 ENN74290.1 ATLV01024452 KE525352 KFB51414.1 AXCN02001183 AAAB01008844 EAA05965.5 NNAY01005290 OXU16782.1 ADMH02001252 ETN63494.1 DS232415 EDS41476.1 GFDL01005412 JAV29633.1 ADMH02000584 ETN65743.1 CH477680 EAT37315.1 CH477340 EAT43109.1 JXUM01061490 JXUM01061491 JXUM01061492 JXUM01061493 KQ562152 KXJ76557.1 JXUM01075103 JXUM01075104 KQ562869 KXJ74964.1 JXUM01120070 KQ566355 KXJ70190.1 AXCM01002234 AXCN02000483 GDIP01175338 JAJ48064.1 APCN01002316 AAAB01008960 EAA10807.3 KQ414612 KOC69187.1 KQ434823 KZC07253.1 GBYB01011296 GBYB01011299 JAG81063.1 JAG81066.1 ATLV01025888 KE525402 KFB52662.1 GFTR01004843 JAW11583.1 LRGB01002992 KZS05223.1 AXCM01003114 DS232003 EDS31344.1 APCN01003158 GFAA01004000 JAT99434.1 KB202640 ESO88740.1 GDIQ01204199 JAK47526.1 GAKP01021363 JAC37589.1
Proteomes
UP000218220
UP000007266
UP000027135
UP000235965
UP000192223
UP000079169
+ More
UP000008237 UP000054359 UP000075809 UP000000311 UP000005203 UP000053097 UP000279307 UP000005205 UP000007755 UP000007819 UP000015103 UP000078542 UP000078492 UP000078541 UP000092461 UP000009046 UP000092462 UP000015104 UP000075884 UP000078540 UP000019118 UP000069272 UP000030765 UP000075903 UP000075886 UP000075880 UP000007062 UP000076407 UP000215335 UP000000673 UP000002320 UP000008820 UP000069940 UP000249989 UP000075881 UP000075883 UP000075885 UP000075900 UP000075902 UP000075840 UP000075882 UP000076408 UP000053825 UP000076502 UP000075920 UP000076858 UP000075901 UP000030746
UP000008237 UP000054359 UP000075809 UP000000311 UP000005203 UP000053097 UP000279307 UP000005205 UP000007755 UP000007819 UP000015103 UP000078542 UP000078492 UP000078541 UP000092461 UP000009046 UP000092462 UP000015104 UP000075884 UP000078540 UP000019118 UP000069272 UP000030765 UP000075903 UP000075886 UP000075880 UP000007062 UP000076407 UP000215335 UP000000673 UP000002320 UP000008820 UP000069940 UP000249989 UP000075881 UP000075883 UP000075885 UP000075900 UP000075902 UP000075840 UP000075882 UP000076408 UP000053825 UP000076502 UP000075920 UP000076858 UP000075901 UP000030746
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JI50
A0A2H1VHX1
A0A2W1BJW2
D6W688
A0A067R802
A0A2J7R1W9
+ More
A0A1B6I5F6 A0A1W4XFI9 A0A1B6MK51 A0A1B6CRF9 A0A1B6EHR9 A0A1Y1NA99 A0A1Y1N739 A0A3Q0J0K6 A0A1B6DTC2 E2BZR8 A0A087TF13 A0A151X8L8 E2A3N4 A0A023F2T9 A0A087ZT40 E9IX31 A0A026WEE3 A0A0C9RRW6 A0A158NPT6 A0A2L2YV77 A0A1Q3FQE7 A0A2R5LD80 F4W7F9 A0A1S4FAF9 J9JRI6 T1HTW9 A0A151I6X6 A0A195EM14 A0A151JT63 A0A1B0CHC5 E0VV64 A0A1B0D9B8 T1KYN9 A0A2H8TR24 A0A182NCM2 A0A195ATE8 A0A336LQ91 N6TY94 A0A182FEA3 A0A084WMH0 A0A182V8Y1 A0A182QVX1 A0A182J744 Q7QG26 A0A182WVD6 A0A232EEP4 W5JL55 B0X6J3 A0A1Q3FQC6 W5JR10 Q16SA0 Q17A55 A0A182GGD2 A0A182GMN1 A0A182H982 A0A182K6H8 A0A182NI78 A0A182M3S7 A0A182PDL6 A0A182R976 A0A182QGX7 A0A182UJX7 A0A182VLX7 A0A0P5D9S1 A0A182I4K8 A0A182X4L1 A0A182LKC2 Q7Q555 A0A182Y2H9 A0A0L7RDX7 A0A154P5X7 A0A0C9R4T5 A0A182WBN8 A0A182JVQ1 A0A182PQQ8 A0A182YJL9 A0A084WR18 A0A224XU05 A0A164MNX4 A0A182LRG3 A0A182W313 A0A182SJE8 A0A182F8F9 A0A182UGW6 B0WMV2 A0A182I917 A0A1E1XJR3 A0A182LAZ8 A0A182J8R6 V4A1D0 A0A0P5JC79 A0A034V4Z1
A0A1B6I5F6 A0A1W4XFI9 A0A1B6MK51 A0A1B6CRF9 A0A1B6EHR9 A0A1Y1NA99 A0A1Y1N739 A0A3Q0J0K6 A0A1B6DTC2 E2BZR8 A0A087TF13 A0A151X8L8 E2A3N4 A0A023F2T9 A0A087ZT40 E9IX31 A0A026WEE3 A0A0C9RRW6 A0A158NPT6 A0A2L2YV77 A0A1Q3FQE7 A0A2R5LD80 F4W7F9 A0A1S4FAF9 J9JRI6 T1HTW9 A0A151I6X6 A0A195EM14 A0A151JT63 A0A1B0CHC5 E0VV64 A0A1B0D9B8 T1KYN9 A0A2H8TR24 A0A182NCM2 A0A195ATE8 A0A336LQ91 N6TY94 A0A182FEA3 A0A084WMH0 A0A182V8Y1 A0A182QVX1 A0A182J744 Q7QG26 A0A182WVD6 A0A232EEP4 W5JL55 B0X6J3 A0A1Q3FQC6 W5JR10 Q16SA0 Q17A55 A0A182GGD2 A0A182GMN1 A0A182H982 A0A182K6H8 A0A182NI78 A0A182M3S7 A0A182PDL6 A0A182R976 A0A182QGX7 A0A182UJX7 A0A182VLX7 A0A0P5D9S1 A0A182I4K8 A0A182X4L1 A0A182LKC2 Q7Q555 A0A182Y2H9 A0A0L7RDX7 A0A154P5X7 A0A0C9R4T5 A0A182WBN8 A0A182JVQ1 A0A182PQQ8 A0A182YJL9 A0A084WR18 A0A224XU05 A0A164MNX4 A0A182LRG3 A0A182W313 A0A182SJE8 A0A182F8F9 A0A182UGW6 B0WMV2 A0A182I917 A0A1E1XJR3 A0A182LAZ8 A0A182J8R6 V4A1D0 A0A0P5JC79 A0A034V4Z1
PDB
6N9G
E-value=1.96298e-50,
Score=500
Ontologies
KEGG
GO
PANTHER
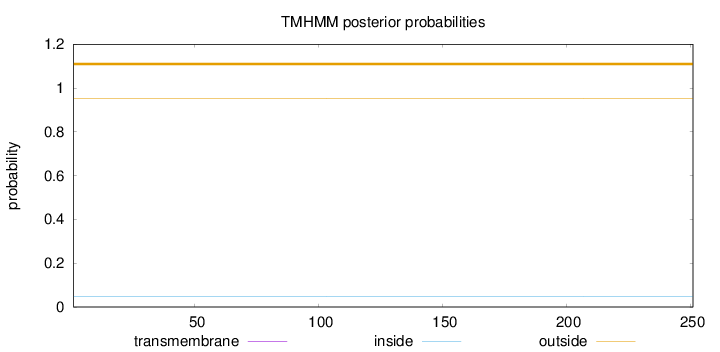
Topology
Subcellular location
Mitochondrion inner membrane
Length:
251
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00037
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.04814
outside
1 - 251
Population Genetic Test Statistics
Pi
121.934657
Theta
125.585306
Tajima's D
-0.515107
CLR
21.876155
CSRT
0.240137993100345
Interpretation
Uncertain
