Gene
KWMTBOMO06281
Pre Gene Modal
BGIBMGA001619
Annotation
PREDICTED:_nucleolysin_TIA-1_isoform_p40_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.773 Nuclear Reliability : 1.345
Sequence
CDS
ATGAACGGGCAGTGGCTCGGCTCCAGGTCCATCCGCACCAACTGGGCCACAAGAAAACCACCGGCTCCGAAGAATGAACTGAACTCCAAGCCTCTGACTTTCGACGAGGTCTACAATCAGAGCTCCGCCAGCAATTGCACGGTGTACTGTGGAGGTCTGACCACCGGCCTCACCGAAGAACTCATGCAGAAGACCTTCCAGCCATTCGGAACTATTCAGGAGATCAGAGTCTTCAAGGACAAAGGATATGCTTTCATCAGATTCTCGACAAAGGAAAGTGCCACCCACGCAATCGTTGCTGTCCACAACACGGATATCAACGGTCAACCAGTCAAATGCTCTTGGGGCAAGGAGTCTGGTGACCCTAACAACTCGCAGGCTCAAGGACAGCTGGGCGGCACAGCTTACAGTCCGTTTGGGACAGCTTACACCGCTAGCGGGGTGCCCCCTGCTTCATACTGGTACAACACGTACCCTCAGCAGATCGGTGGCTTCCTGCAGGGCGTCCAAGGCGTGCAGGGGTACACTTACGCTGGACAGTTTGGATATCAACAGCAGTACATGGGGTGA
Protein
MNGQWLGSRSIRTNWATRKPPAPKNELNSKPLTFDEVYNQSSASNCTVYCGGLTTGLTEELMQKTFQPFGTIQEIRVFKDKGYAFIRFSTKESATHAIVAVHNTDINGQPVKCSWGKESGDPNNSQAQGQLGGTAYSPFGTAYTASGVPPASYWYNTYPQQIGGFLQGVQGVQGYTYAGQFGYQQQYMG
Summary
Uniprot
H9IWI7
A0A194Q5W6
A0A0N0PEN8
A0A2A4JRJ0
A0A2W1BPB5
A0A2H1VJX4
+ More
A0A1B6E4A1 A0A067RIJ2 A0A0A9XME9 A0A232FLG1 A0A087ZTY2 A0A023F4M2 A0A154PLH0 A0A3L8DYK6 A0A151WKF0 A0A0M9A301 A0A1Y1L3L3 K7IUA4 A0A158NYN1 A0A195CE14 A0A212F5Q8 A0A195BRI5 A0A2S2QGR1 A0A2H8TL97 A0A195DDD4 A0A2S2N8S4 N6UHC7 J9JRG5 U4TUK0 A0A2S2QKD0 A0A139WP22 A0A1B0CI93 A0A3R7M7K7 A0A182JHS4 A0A2P2IF94 Q16WX1 A0A182WK46 A0A182N242 A0A182ICX8 A0A182PQ65 A0A084VXL5 A0A182RQD3 Q7PSZ0 W8C6W1 A0A1S4FL99 A0A182YP10 A0A182WRZ2 W5JHL8 B4IGZ6 A4IJ77 A0A1W4U548 B4JTL1 A0A3B0KCL0 B4QY71 B4K8Q2 B4LYK5 A0A0M4EW15 B4G5U4 B3M0H7 E1JJ00 B3P5U2 A0A126GV08 B4PR64 A0A2R5LKK2 B4NFH7 A0A0Q9X775 A0A1I8NXU1 A0A0H4Y1N6 A8JPX0 A0A131XWL8 A0A3B0K8M2 A0A3B0JM18 B4G6C3 A0A1J1I5A0 B5DWJ8 A0A1I8MKD9 A0A3B0K0P7 A0A0R3NG74 A0A023FLE7
A0A1B6E4A1 A0A067RIJ2 A0A0A9XME9 A0A232FLG1 A0A087ZTY2 A0A023F4M2 A0A154PLH0 A0A3L8DYK6 A0A151WKF0 A0A0M9A301 A0A1Y1L3L3 K7IUA4 A0A158NYN1 A0A195CE14 A0A212F5Q8 A0A195BRI5 A0A2S2QGR1 A0A2H8TL97 A0A195DDD4 A0A2S2N8S4 N6UHC7 J9JRG5 U4TUK0 A0A2S2QKD0 A0A139WP22 A0A1B0CI93 A0A3R7M7K7 A0A182JHS4 A0A2P2IF94 Q16WX1 A0A182WK46 A0A182N242 A0A182ICX8 A0A182PQ65 A0A084VXL5 A0A182RQD3 Q7PSZ0 W8C6W1 A0A1S4FL99 A0A182YP10 A0A182WRZ2 W5JHL8 B4IGZ6 A4IJ77 A0A1W4U548 B4JTL1 A0A3B0KCL0 B4QY71 B4K8Q2 B4LYK5 A0A0M4EW15 B4G5U4 B3M0H7 E1JJ00 B3P5U2 A0A126GV08 B4PR64 A0A2R5LKK2 B4NFH7 A0A0Q9X775 A0A1I8NXU1 A0A0H4Y1N6 A8JPX0 A0A131XWL8 A0A3B0K8M2 A0A3B0JM18 B4G6C3 A0A1J1I5A0 B5DWJ8 A0A1I8MKD9 A0A3B0K0P7 A0A0R3NG74 A0A023FLE7
Pubmed
19121390
26354079
28756777
24845553
25401762
28648823
+ More
25474469 30249741 28004739 20075255 21347285 22118469 23537049 18362917 19820115 17510324 24438588 12364791 24495485 25244985 20920257 23761445 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 25315136
25474469 30249741 28004739 20075255 21347285 22118469 23537049 18362917 19820115 17510324 24438588 12364791 24495485 25244985 20920257 23761445 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 25315136
EMBL
BABH01021300
BABH01021301
BABH01021302
KQ459463
KPJ00400.1
KQ459891
+ More
KPJ19547.1 NWSH01000758 PCG74386.1 KZ150008 PZC75147.1 ODYU01002975 SOQ41149.1 GEDC01004546 JAS32752.1 KK852661 KDR19108.1 GBHO01023616 JAG19988.1 NNAY01000054 OXU31492.1 GBBI01002420 JAC16292.1 KQ434968 KZC12706.1 QOIP01000002 RLU25372.1 KQ983012 KYQ48290.1 KQ435752 KOX76177.1 GEZM01067848 GEZM01067847 JAV67381.1 AAZX01002623 AAZX01002629 AAZX01004673 AAZX01006610 AAZX01006771 AAZX01012594 AAZX01013265 AAZX01015850 AAZX01018968 AAZX01021525 ADTU01000687 ADTU01000688 ADTU01000689 ADTU01000690 ADTU01000691 ADTU01000692 ADTU01000693 ADTU01000694 ADTU01000695 ADTU01000696 KQ977876 KYM99052.1 AGBW02010146 OWR49072.1 KQ976423 KYM88829.1 GGMS01007159 MBY76362.1 GFXV01003109 MBW14914.1 KQ980979 KYN10837.1 GGMR01000936 MBY13555.1 APGK01020359 KB740193 ENN81120.1 ABLF02040599 ABLF02040601 ABLF02040605 ABLF02040618 ABLF02040619 ABLF02040620 ABLF02040622 KB631669 ERL85234.1 GGMS01008994 MBY78197.1 KQ971307 KYB29607.1 AJWK01013111 QCYY01001922 ROT74223.1 IACF01007047 LAB72630.1 CH477552 EAT39097.1 APCN01006061 APCN01006062 ATLV01018097 KE525212 KFB42709.1 AAAB01008810 EAA04819.5 GAMC01008316 JAB98239.1 ADMH02001370 ETN62803.1 CH480837 EDW49114.1 BT030435 ABO52855.1 CH916374 EDV91440.1 OUUW01000005 SPP81368.1 CM000364 EDX14719.1 CH933806 EDW15471.2 CH940650 EDW68025.2 CP012526 ALC47642.1 CH479179 EDW23938.1 CH902617 EDV44224.2 AE014297 ACZ95049.2 ACZ95050.2 CH954182 EDV53342.1 ALI30647.1 CM000160 EDW98428.2 GGLE01005916 MBY10042.1 CH964251 EDW83044.2 KRG01578.1 BT150486 AKR06214.1 ABW08789.2 GEFM01004347 JAP71449.1 SPP81371.1 SPP81372.1 EDW23943.1 CVRI01000040 CRK94764.1 CM000070 EDY67785.2 SPP81370.1 KRS99994.1 GBBK01002994 JAC21488.1
KPJ19547.1 NWSH01000758 PCG74386.1 KZ150008 PZC75147.1 ODYU01002975 SOQ41149.1 GEDC01004546 JAS32752.1 KK852661 KDR19108.1 GBHO01023616 JAG19988.1 NNAY01000054 OXU31492.1 GBBI01002420 JAC16292.1 KQ434968 KZC12706.1 QOIP01000002 RLU25372.1 KQ983012 KYQ48290.1 KQ435752 KOX76177.1 GEZM01067848 GEZM01067847 JAV67381.1 AAZX01002623 AAZX01002629 AAZX01004673 AAZX01006610 AAZX01006771 AAZX01012594 AAZX01013265 AAZX01015850 AAZX01018968 AAZX01021525 ADTU01000687 ADTU01000688 ADTU01000689 ADTU01000690 ADTU01000691 ADTU01000692 ADTU01000693 ADTU01000694 ADTU01000695 ADTU01000696 KQ977876 KYM99052.1 AGBW02010146 OWR49072.1 KQ976423 KYM88829.1 GGMS01007159 MBY76362.1 GFXV01003109 MBW14914.1 KQ980979 KYN10837.1 GGMR01000936 MBY13555.1 APGK01020359 KB740193 ENN81120.1 ABLF02040599 ABLF02040601 ABLF02040605 ABLF02040618 ABLF02040619 ABLF02040620 ABLF02040622 KB631669 ERL85234.1 GGMS01008994 MBY78197.1 KQ971307 KYB29607.1 AJWK01013111 QCYY01001922 ROT74223.1 IACF01007047 LAB72630.1 CH477552 EAT39097.1 APCN01006061 APCN01006062 ATLV01018097 KE525212 KFB42709.1 AAAB01008810 EAA04819.5 GAMC01008316 JAB98239.1 ADMH02001370 ETN62803.1 CH480837 EDW49114.1 BT030435 ABO52855.1 CH916374 EDV91440.1 OUUW01000005 SPP81368.1 CM000364 EDX14719.1 CH933806 EDW15471.2 CH940650 EDW68025.2 CP012526 ALC47642.1 CH479179 EDW23938.1 CH902617 EDV44224.2 AE014297 ACZ95049.2 ACZ95050.2 CH954182 EDV53342.1 ALI30647.1 CM000160 EDW98428.2 GGLE01005916 MBY10042.1 CH964251 EDW83044.2 KRG01578.1 BT150486 AKR06214.1 ABW08789.2 GEFM01004347 JAP71449.1 SPP81371.1 SPP81372.1 EDW23943.1 CVRI01000040 CRK94764.1 CM000070 EDY67785.2 SPP81370.1 KRS99994.1 GBBK01002994 JAC21488.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000027135
UP000215335
+ More
UP000005203 UP000076502 UP000279307 UP000075809 UP000053105 UP000002358 UP000005205 UP000078542 UP000007151 UP000078540 UP000078492 UP000019118 UP000007819 UP000030742 UP000007266 UP000092461 UP000283509 UP000075880 UP000008820 UP000075920 UP000075884 UP000075840 UP000075885 UP000030765 UP000075900 UP000007062 UP000076408 UP000076407 UP000000673 UP000001292 UP000192221 UP000001070 UP000268350 UP000000304 UP000009192 UP000008792 UP000092553 UP000008744 UP000007801 UP000000803 UP000008711 UP000002282 UP000007798 UP000095300 UP000183832 UP000001819 UP000095301
UP000005203 UP000076502 UP000279307 UP000075809 UP000053105 UP000002358 UP000005205 UP000078542 UP000007151 UP000078540 UP000078492 UP000019118 UP000007819 UP000030742 UP000007266 UP000092461 UP000283509 UP000075880 UP000008820 UP000075920 UP000075884 UP000075840 UP000075885 UP000030765 UP000075900 UP000007062 UP000076408 UP000076407 UP000000673 UP000001292 UP000192221 UP000001070 UP000268350 UP000000304 UP000009192 UP000008792 UP000092553 UP000008744 UP000007801 UP000000803 UP000008711 UP000002282 UP000007798 UP000095300 UP000183832 UP000001819 UP000095301
PRIDE
Pfam
PF00076 RRM_1
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
ProteinModelPortal
H9IWI7
A0A194Q5W6
A0A0N0PEN8
A0A2A4JRJ0
A0A2W1BPB5
A0A2H1VJX4
+ More
A0A1B6E4A1 A0A067RIJ2 A0A0A9XME9 A0A232FLG1 A0A087ZTY2 A0A023F4M2 A0A154PLH0 A0A3L8DYK6 A0A151WKF0 A0A0M9A301 A0A1Y1L3L3 K7IUA4 A0A158NYN1 A0A195CE14 A0A212F5Q8 A0A195BRI5 A0A2S2QGR1 A0A2H8TL97 A0A195DDD4 A0A2S2N8S4 N6UHC7 J9JRG5 U4TUK0 A0A2S2QKD0 A0A139WP22 A0A1B0CI93 A0A3R7M7K7 A0A182JHS4 A0A2P2IF94 Q16WX1 A0A182WK46 A0A182N242 A0A182ICX8 A0A182PQ65 A0A084VXL5 A0A182RQD3 Q7PSZ0 W8C6W1 A0A1S4FL99 A0A182YP10 A0A182WRZ2 W5JHL8 B4IGZ6 A4IJ77 A0A1W4U548 B4JTL1 A0A3B0KCL0 B4QY71 B4K8Q2 B4LYK5 A0A0M4EW15 B4G5U4 B3M0H7 E1JJ00 B3P5U2 A0A126GV08 B4PR64 A0A2R5LKK2 B4NFH7 A0A0Q9X775 A0A1I8NXU1 A0A0H4Y1N6 A8JPX0 A0A131XWL8 A0A3B0K8M2 A0A3B0JM18 B4G6C3 A0A1J1I5A0 B5DWJ8 A0A1I8MKD9 A0A3B0K0P7 A0A0R3NG74 A0A023FLE7
A0A1B6E4A1 A0A067RIJ2 A0A0A9XME9 A0A232FLG1 A0A087ZTY2 A0A023F4M2 A0A154PLH0 A0A3L8DYK6 A0A151WKF0 A0A0M9A301 A0A1Y1L3L3 K7IUA4 A0A158NYN1 A0A195CE14 A0A212F5Q8 A0A195BRI5 A0A2S2QGR1 A0A2H8TL97 A0A195DDD4 A0A2S2N8S4 N6UHC7 J9JRG5 U4TUK0 A0A2S2QKD0 A0A139WP22 A0A1B0CI93 A0A3R7M7K7 A0A182JHS4 A0A2P2IF94 Q16WX1 A0A182WK46 A0A182N242 A0A182ICX8 A0A182PQ65 A0A084VXL5 A0A182RQD3 Q7PSZ0 W8C6W1 A0A1S4FL99 A0A182YP10 A0A182WRZ2 W5JHL8 B4IGZ6 A4IJ77 A0A1W4U548 B4JTL1 A0A3B0KCL0 B4QY71 B4K8Q2 B4LYK5 A0A0M4EW15 B4G5U4 B3M0H7 E1JJ00 B3P5U2 A0A126GV08 B4PR64 A0A2R5LKK2 B4NFH7 A0A0Q9X775 A0A1I8NXU1 A0A0H4Y1N6 A8JPX0 A0A131XWL8 A0A3B0K8M2 A0A3B0JM18 B4G6C3 A0A1J1I5A0 B5DWJ8 A0A1I8MKD9 A0A3B0K0P7 A0A0R3NG74 A0A023FLE7
PDB
2MJN
E-value=1.73657e-42,
Score=430
Ontologies
GO
Topology
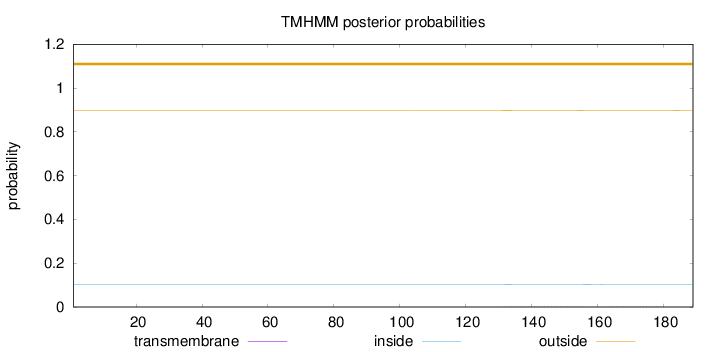
Length:
189
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08405
Exp number, first 60 AAs:
0.00289
Total prob of N-in:
0.10280
outside
1 - 189
Population Genetic Test Statistics
Pi
169.224942
Theta
163.187094
Tajima's D
0.768672
CLR
0.460528
CSRT
0.60121993900305
Interpretation
Uncertain
