Gene
KWMTBOMO06279 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011051
Annotation
PREDICTED:_uncharacterized_protein_LOC105841272_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.142 Nuclear Reliability : 1.74
Sequence
CDS
ATGCACGTTAATTATATATTAACATTTCTCTTCGCCTGGAACTTAACGATTTTGACCAGCAGCCAGTTCGTTGGTTTTGTGCCGTCAAATATAGTTGAGAATCAGCCGCCAATCACAAATAGGCCCGTCACATATTCGCCTGGCCCTGTCGATAGCTTTCAAGAAGATGAACCGACGTTGGAGATAGGTCCTGATGAAGACGACATGACTTCGGTGGAGAAGAGGCGATACTTTGAAATTGCTGAGCGAATGTGCGATAAATACAAATCGTTGAACACAAAAAAGATAGTGGCCATCCCACTGCTTCCGTCGGCGCATGGCGTCGAAGTAAATGTCTCGGATTGTACACCCATCAAAATACCGCTTATCATCGGAGGGCAGGTCGCTACGATCGAGGAGTTCCCGCACATGGCCTTAATAGGATGGAAGAATGCACAGGACTATGGATATTCTTGGAAGTGCGGGGGGTCGCTCATCTGCGACGAATACGTCCTGACTGCCGGTCATTGCACGTACCAGGAAAAGGATTACAGCGTCATCAGCGGAACACCCCGCGCTGTTCAACTCGGGTCGGCGCGCCGCGACGACCGCCGCGCGTTGATCCTCGGCATCTCCACCGTCATACGACACCCGAAGTACAAACGGAGGAGGTCCTACTACGACTTGGCTCTTGTTAAGCTGGTCTCCAAAGTGAGATTCTCGGACCAGATCAGGCCGGCTTGTCTAGGAGTTCCACCGGCAGAAGGGGAGAGAGTCGTCGTCAGCGGCTGGGGACGAACCGAATTTGGTGGAGACGCATCTGCGGACCTTCGGAGCGTTTCCGTTGAAATCTGGAATATGCAAGAGTGCAGGGAGACATGGGGCACCTCGTTGAAGCTGCCAAACGGACCCACGCCTGATAGCCACGTCTGCGCTGGTGAGAGGAAAGGGGGCAAGGACACATGTCAGGGGGACTCCGGTGGGCCAGCCCAGGTGGAAGACGGCTGCGTGTGGCGCGTCGTGGCCGTCACCTCGATCGGGAGGTCCTGCGCCGCTCCGCAGACTCCAGCGCTATACTCCAGATTACATATACCGTTTATAATCGCACAAATATTCAACAACCAGCCGTCCAACAACAATTTTAATGGTCAAACAATCGACGATCGGAATCAATATTACAATCAAAACGTCAACAATCAAAACAAGCACACCCAGAACAGTAATAATCAAGTTTTATTTAATCAAGATCATTATAATGCTCCGAGTTATGATGCCCAGGATCATTACAGCAGCCAGGATAATCAGTATAACAATCAGTACCAACAGGATGTAAGACGTCCGGAGGATACATGCTGA
Protein
MHVNYILTFLFAWNLTILTSSQFVGFVPSNIVENQPPITNRPVTYSPGPVDSFQEDEPTLEIGPDEDDMTSVEKRRYFEIAERMCDKYKSLNTKKIVAIPLLPSAHGVEVNVSDCTPIKIPLIIGGQVATIEEFPHMALIGWKNAQDYGYSWKCGGSLICDEYVLTAGHCTYQEKDYSVISGTPRAVQLGSARRDDRRALILGISTVIRHPKYKRRRSYYDLALVKLVSKVRFSDQIRPACLGVPPAEGERVVVSGWGRTEFGGDASADLRSVSVEIWNMQECRETWGTSLKLPNGPTPDSHVCAGERKGGKDTCQGDSGGPAQVEDGCVWRVVAVTSIGRSCAAPQTPALYSRLHIPFIIAQIFNNQPSNNNFNGQTIDDRNQYYNQNVNNQNKHTQNSNNQVLFNQDHYNAPSYDAQDHYSSQDNQYNNQYQQDVRRPEDTC
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
2F83
E-value=2.91062e-28,
Score=312
Ontologies
GO
Topology
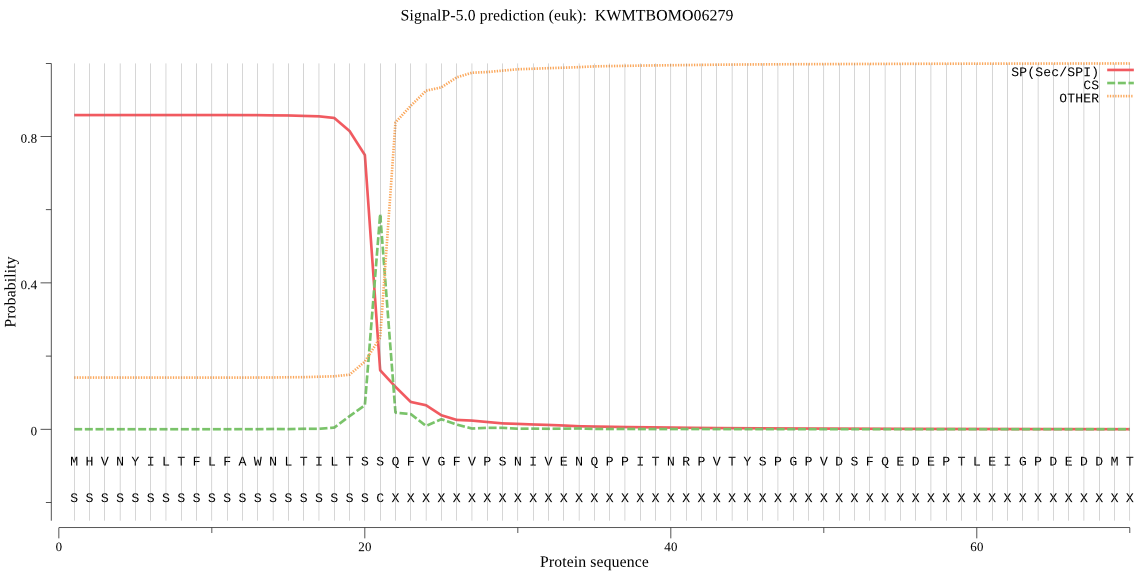
SignalP
Position: 1 - 21,
Likelihood: 0.858402
Length:
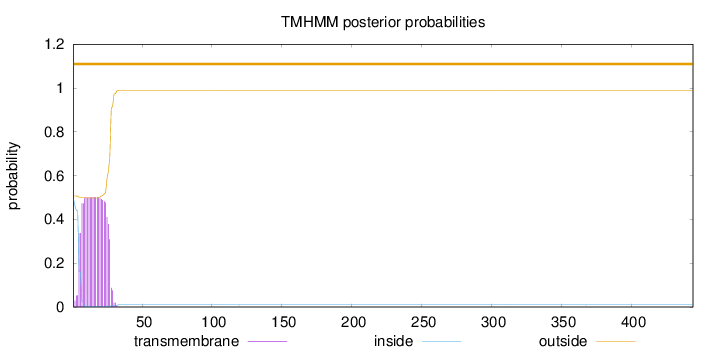
444
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.00754
Exp number, first 60 AAs:
11.00209
Total prob of N-in:
0.49322
POSSIBLE N-term signal
sequence
outside
1 - 444
Population Genetic Test Statistics
Pi
229.89144
Theta
177.66458
Tajima's D
0.578213
CLR
180.82549
CSRT
0.542322883855807
Interpretation
Uncertain
