Gene
KWMTBOMO06277
Annotation
PREDICTED:_ras-related_protein_Rab-24-like_[Plutella_xylostella]
Full name
Ras-related protein Rab-24
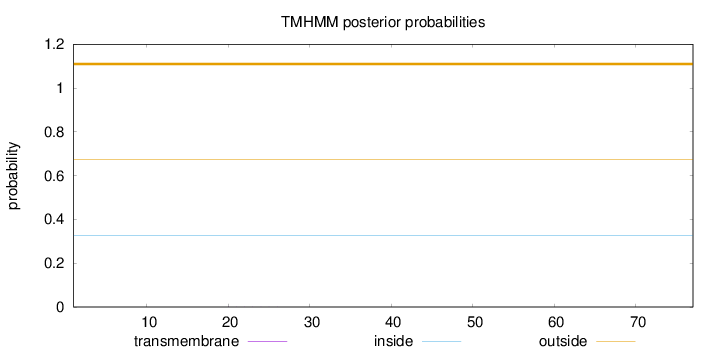
Location in the cell
Extracellular Reliability : 1.563
Sequence
CDS
ATGAGTCGATGTGATTTTAAAATAGTGCTTTTAGGCAGCGAACACGTCGGAAAGACGAGCTTAGTTTTAAGATTCGTGAATTGTAGGTTCAACGCTGAGATACCATATCAAAATACCGTCGGTGCTGCATTTTGCGCTAAAGCTATGCGATCAAAAGAAAAAGACTACAGTATTGGCATCTGGGATACAGCAGGCAGCGAAAGGTATATTAATAATTATGTACATTGTATATAG
Protein
MSRCDFKIVLLGSEHVGKTSLVLRFVNCRFNAEIPYQNTVGAAFCAKAMRSKEKDYSIGIWDTAGSERYINNYVHCI
Summary
Description
May be involved in autophagy-related processes.
Subunit
Unlike other Rab family members, does not interact with GDP dissociation inhibitors (GDIs), including ARHGDIA and ARHGDIB. Interacts with ZFYVE20.
Miscellaneous
The unusual Ser-67, instead of a conserved Gln in other family members, is the cause of low GTPase activity. As a result, the predominant nucleotide associated with the protein is GTP (By similarity).
Similarity
Belongs to the small GTPase superfamily. Rab family.
Keywords
Autophagy
Complete proteome
Cytoplasm
GTP-binding
Lipoprotein
Membrane
Nucleotide-binding
Prenylation
Protein transport
Reference proteome
Transport
Feature
chain Ras-related protein Rab-24
Uniprot
A0A194Q4D6
A0A0N0PEA2
A0A2H1V5D4
A0A2A4JQQ2
A0A1E1WRL3
A0A212F0Y1
+ More
A0A2J7PQN6 A0A2P8Y430 K7J6I0 A0A232ESN9 A0A0M8ZZ71 A0A2A3EER8 A0A088A4K1 A0A154PKK7 T2M8J9 A0A2Y9QSA9 A0A2Y9EJM7 E1B771 A0A3Q1M6P8 L9JBN8 A0A0L7QMM0 A0A1A6GHY1 C3Y491 A0A061IBW5 A0A067RHX9 A0A1Z5KWR4 W4Z213 A0A2R5L465 L7LUG3 A0A023FUW3 A0A293LBI9 A0A224Z5J3 A0A131Z646 L7M734 A0A224Z7H1 G3ML32 A0A023GFU7 A0A1E1WZF1 A0A131XJP9 A0A2B4RZB2 A0A026WAD7 A0A2Y9QLB7 A0A2Y9QFV8 A0A0K8R8Z2 A0A3Q7TLT2 A0A2Y9HZA2 A0A2U3ZGP9 A0A3Q7U4M6 U6DW12 A0A3Q7NN36 A0A2Y9HZQ3 A0A091DUX7 F6V5H3 A0A384C7V0 A0A3Q7NNG3 A0A2Y9HZA6 A0A3Q7WY86 L5JV16 A0A384C6T0 A0A3Q7XUM0 G1TK72 A0A1S3IJZ8 H0XQF3 D2HZ82 B7PLK7 V9L127 A0A250XW42 A0A2U3XPB0 G1Q6N8 K9IWQ8 A0A2K5RMM0 Q3T107 A0A2K6ARW3 A0A2K6LR30 A0A384C6V1 G1RQA7 A0A2K5YTH6 A0A0D9R986 A0A2R9BW20 A0A096MWW9 A0A2K5MWH9 A0A2U3W0R9 A0A2K5I1Z0 A0A3Q7W102 G7P700 G3RS69 H2QS35 G7MU36 Q969Q5 I3MWQ8 E2R4M1 F6XZH4 A0A2Y9DBD6 A0A3Q7NWZ5 A0A2K5CR06 A0A2Y9HZ97 A0A2U3XP12 A0A2I2UYV4 L5LY47 A0A3Q7T1R8
A0A2J7PQN6 A0A2P8Y430 K7J6I0 A0A232ESN9 A0A0M8ZZ71 A0A2A3EER8 A0A088A4K1 A0A154PKK7 T2M8J9 A0A2Y9QSA9 A0A2Y9EJM7 E1B771 A0A3Q1M6P8 L9JBN8 A0A0L7QMM0 A0A1A6GHY1 C3Y491 A0A061IBW5 A0A067RHX9 A0A1Z5KWR4 W4Z213 A0A2R5L465 L7LUG3 A0A023FUW3 A0A293LBI9 A0A224Z5J3 A0A131Z646 L7M734 A0A224Z7H1 G3ML32 A0A023GFU7 A0A1E1WZF1 A0A131XJP9 A0A2B4RZB2 A0A026WAD7 A0A2Y9QLB7 A0A2Y9QFV8 A0A0K8R8Z2 A0A3Q7TLT2 A0A2Y9HZA2 A0A2U3ZGP9 A0A3Q7U4M6 U6DW12 A0A3Q7NN36 A0A2Y9HZQ3 A0A091DUX7 F6V5H3 A0A384C7V0 A0A3Q7NNG3 A0A2Y9HZA6 A0A3Q7WY86 L5JV16 A0A384C6T0 A0A3Q7XUM0 G1TK72 A0A1S3IJZ8 H0XQF3 D2HZ82 B7PLK7 V9L127 A0A250XW42 A0A2U3XPB0 G1Q6N8 K9IWQ8 A0A2K5RMM0 Q3T107 A0A2K6ARW3 A0A2K6LR30 A0A384C6V1 G1RQA7 A0A2K5YTH6 A0A0D9R986 A0A2R9BW20 A0A096MWW9 A0A2K5MWH9 A0A2U3W0R9 A0A2K5I1Z0 A0A3Q7W102 G7P700 G3RS69 H2QS35 G7MU36 Q969Q5 I3MWQ8 E2R4M1 F6XZH4 A0A2Y9DBD6 A0A3Q7NWZ5 A0A2K5CR06 A0A2Y9HZ97 A0A2U3XP12 A0A2I2UYV4 L5LY47 A0A3Q7T1R8
Pubmed
26354079
22118469
29403074
20075255
28648823
24065732
+ More
19393038 23385571 18563158 23929341 24845553 28528879 25576852 28797301 26830274 22216098 28503490 28049606 24508170 30249741 17495919 23258410 24813606 21993624 20010809 24402279 28087693 22722832 22002653 22398555 16136131 17431167 25319552 15489334 17974005 10660536 16034420 21269460 16341006 19892987 17975172
19393038 23385571 18563158 23929341 24845553 28528879 25576852 28797301 26830274 22216098 28503490 28049606 24508170 30249741 17495919 23258410 24813606 21993624 20010809 24402279 28087693 22722832 22002653 22398555 16136131 17431167 25319552 15489334 17974005 10660536 16034420 21269460 16341006 19892987 17975172
EMBL
KQ459463
KPJ00403.1
KQ459891
KPJ19549.1
ODYU01000740
SOQ35986.1
+ More
NWSH01000758 PCG74385.1 GDQN01001613 JAT89441.1 AGBW02011017 OWR47364.1 NEVH01022635 PNF18642.1 PYGN01000952 PSN39005.1 NNAY01002393 OXU21371.1 KQ435803 KOX73086.1 KZ288266 PBC30255.1 KQ434948 KZC12399.1 HAAD01002028 CDG68260.1 KB321078 ELW47980.1 KQ414885 KOC59873.1 LZPO01096785 OBS65479.1 GG666484 EEN65117.1 KE671212 ERE80482.1 KK852669 KDR18847.1 GFJQ02007408 JAV99561.1 AAGJ04055392 AAGJ04055393 AAGJ04055394 GGLE01000162 MBY04288.1 GACK01010550 JAA54484.1 GBBL01002041 JAC25279.1 GFWV01000421 MAA25151.1 GFPF01011225 MAA22371.1 GEDV01002295 JAP86262.1 GACK01006066 JAA58968.1 GFPF01011224 MAA22370.1 JO842583 AEO34200.1 GBBM01002641 JAC32777.1 GFAC01006788 JAT92400.1 GEFH01002231 JAP66350.1 LSMT01000196 PFX23794.1 KK107323 QOIP01000002 EZA52596.1 RLU25573.1 GADI01006455 JAA67353.1 HAAF01014859 CCP86681.1 KN122016 KFO34090.1 KB031114 ELK03150.1 AAGW02066612 AAQR03181150 ACTA01043487 GL193783 EFB17690.1 ABJB010113239 ABJB010182785 ABJB010332394 ABJB010489088 ABJB010645540 ABJB010782999 ABJB010924951 ABJB010970624 ABJB011038339 ABJB011073917 DS740966 EEC07479.1 JW872065 AFP04583.1 GFFV01004352 JAV35593.1 AAPE02001259 GABZ01007979 JAA45546.1 BC102183 AAI02184.1 ADFV01124990 AQIB01152928 AJFE02019372 AHZZ02030477 AQIA01061548 CM001281 EHH54796.1 CABD030042611 AACZ04060243 GABC01001087 GABC01001086 GABF01001862 GABF01001861 GABD01008146 GABD01008145 GABE01010897 GABE01010896 NBAG03000516 JAA10251.1 JAA20283.1 JAA24954.1 JAA33842.1 PNI18094.1 PNI18095.1 JSUE03037611 JU319791 JU472881 JU476042 JU476043 JV047403 JV047404 CM001258 AFE63547.1 AFH29685.1 AFI37474.1 EHH27076.1 AF087904 BT007268 CH471195 BC010006 BC015534 BC021263 AL833898 AGTP01071708 AAEX03002967 AANG04001174 KB106582 ELK30950.1
NWSH01000758 PCG74385.1 GDQN01001613 JAT89441.1 AGBW02011017 OWR47364.1 NEVH01022635 PNF18642.1 PYGN01000952 PSN39005.1 NNAY01002393 OXU21371.1 KQ435803 KOX73086.1 KZ288266 PBC30255.1 KQ434948 KZC12399.1 HAAD01002028 CDG68260.1 KB321078 ELW47980.1 KQ414885 KOC59873.1 LZPO01096785 OBS65479.1 GG666484 EEN65117.1 KE671212 ERE80482.1 KK852669 KDR18847.1 GFJQ02007408 JAV99561.1 AAGJ04055392 AAGJ04055393 AAGJ04055394 GGLE01000162 MBY04288.1 GACK01010550 JAA54484.1 GBBL01002041 JAC25279.1 GFWV01000421 MAA25151.1 GFPF01011225 MAA22371.1 GEDV01002295 JAP86262.1 GACK01006066 JAA58968.1 GFPF01011224 MAA22370.1 JO842583 AEO34200.1 GBBM01002641 JAC32777.1 GFAC01006788 JAT92400.1 GEFH01002231 JAP66350.1 LSMT01000196 PFX23794.1 KK107323 QOIP01000002 EZA52596.1 RLU25573.1 GADI01006455 JAA67353.1 HAAF01014859 CCP86681.1 KN122016 KFO34090.1 KB031114 ELK03150.1 AAGW02066612 AAQR03181150 ACTA01043487 GL193783 EFB17690.1 ABJB010113239 ABJB010182785 ABJB010332394 ABJB010489088 ABJB010645540 ABJB010782999 ABJB010924951 ABJB010970624 ABJB011038339 ABJB011073917 DS740966 EEC07479.1 JW872065 AFP04583.1 GFFV01004352 JAV35593.1 AAPE02001259 GABZ01007979 JAA45546.1 BC102183 AAI02184.1 ADFV01124990 AQIB01152928 AJFE02019372 AHZZ02030477 AQIA01061548 CM001281 EHH54796.1 CABD030042611 AACZ04060243 GABC01001087 GABC01001086 GABF01001862 GABF01001861 GABD01008146 GABD01008145 GABE01010897 GABE01010896 NBAG03000516 JAA10251.1 JAA20283.1 JAA24954.1 JAA33842.1 PNI18094.1 PNI18095.1 JSUE03037611 JU319791 JU472881 JU476042 JU476043 JV047403 JV047404 CM001258 AFE63547.1 AFH29685.1 AFI37474.1 EHH27076.1 AF087904 BT007268 CH471195 BC010006 BC015534 BC021263 AL833898 AGTP01071708 AAEX03002967 AANG04001174 KB106582 ELK30950.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000007151
UP000235965
UP000245037
+ More
UP000002358 UP000215335 UP000053105 UP000242457 UP000005203 UP000076502 UP000248480 UP000248484 UP000009136 UP000011518 UP000053825 UP000092124 UP000001554 UP000030759 UP000027135 UP000007110 UP000225706 UP000053097 UP000279307 UP000286640 UP000248481 UP000245340 UP000286641 UP000028990 UP000002280 UP000261680 UP000286642 UP000010552 UP000291021 UP000001811 UP000085678 UP000005225 UP000008912 UP000001555 UP000245341 UP000001074 UP000233040 UP000233120 UP000233180 UP000001073 UP000233140 UP000029965 UP000240080 UP000028761 UP000233060 UP000233080 UP000009130 UP000233100 UP000001519 UP000002277 UP000006718 UP000005640 UP000005215 UP000002254 UP000002281 UP000233020 UP000011712
UP000002358 UP000215335 UP000053105 UP000242457 UP000005203 UP000076502 UP000248480 UP000248484 UP000009136 UP000011518 UP000053825 UP000092124 UP000001554 UP000030759 UP000027135 UP000007110 UP000225706 UP000053097 UP000279307 UP000286640 UP000248481 UP000245340 UP000286641 UP000028990 UP000002280 UP000261680 UP000286642 UP000010552 UP000291021 UP000001811 UP000085678 UP000005225 UP000008912 UP000001555 UP000245341 UP000001074 UP000233040 UP000233120 UP000233180 UP000001073 UP000233140 UP000029965 UP000240080 UP000028761 UP000233060 UP000233080 UP000009130 UP000233100 UP000001519 UP000002277 UP000006718 UP000005640 UP000005215 UP000002254 UP000002281 UP000233020 UP000011712
PRIDE
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A194Q4D6
A0A0N0PEA2
A0A2H1V5D4
A0A2A4JQQ2
A0A1E1WRL3
A0A212F0Y1
+ More
A0A2J7PQN6 A0A2P8Y430 K7J6I0 A0A232ESN9 A0A0M8ZZ71 A0A2A3EER8 A0A088A4K1 A0A154PKK7 T2M8J9 A0A2Y9QSA9 A0A2Y9EJM7 E1B771 A0A3Q1M6P8 L9JBN8 A0A0L7QMM0 A0A1A6GHY1 C3Y491 A0A061IBW5 A0A067RHX9 A0A1Z5KWR4 W4Z213 A0A2R5L465 L7LUG3 A0A023FUW3 A0A293LBI9 A0A224Z5J3 A0A131Z646 L7M734 A0A224Z7H1 G3ML32 A0A023GFU7 A0A1E1WZF1 A0A131XJP9 A0A2B4RZB2 A0A026WAD7 A0A2Y9QLB7 A0A2Y9QFV8 A0A0K8R8Z2 A0A3Q7TLT2 A0A2Y9HZA2 A0A2U3ZGP9 A0A3Q7U4M6 U6DW12 A0A3Q7NN36 A0A2Y9HZQ3 A0A091DUX7 F6V5H3 A0A384C7V0 A0A3Q7NNG3 A0A2Y9HZA6 A0A3Q7WY86 L5JV16 A0A384C6T0 A0A3Q7XUM0 G1TK72 A0A1S3IJZ8 H0XQF3 D2HZ82 B7PLK7 V9L127 A0A250XW42 A0A2U3XPB0 G1Q6N8 K9IWQ8 A0A2K5RMM0 Q3T107 A0A2K6ARW3 A0A2K6LR30 A0A384C6V1 G1RQA7 A0A2K5YTH6 A0A0D9R986 A0A2R9BW20 A0A096MWW9 A0A2K5MWH9 A0A2U3W0R9 A0A2K5I1Z0 A0A3Q7W102 G7P700 G3RS69 H2QS35 G7MU36 Q969Q5 I3MWQ8 E2R4M1 F6XZH4 A0A2Y9DBD6 A0A3Q7NWZ5 A0A2K5CR06 A0A2Y9HZ97 A0A2U3XP12 A0A2I2UYV4 L5LY47 A0A3Q7T1R8
A0A2J7PQN6 A0A2P8Y430 K7J6I0 A0A232ESN9 A0A0M8ZZ71 A0A2A3EER8 A0A088A4K1 A0A154PKK7 T2M8J9 A0A2Y9QSA9 A0A2Y9EJM7 E1B771 A0A3Q1M6P8 L9JBN8 A0A0L7QMM0 A0A1A6GHY1 C3Y491 A0A061IBW5 A0A067RHX9 A0A1Z5KWR4 W4Z213 A0A2R5L465 L7LUG3 A0A023FUW3 A0A293LBI9 A0A224Z5J3 A0A131Z646 L7M734 A0A224Z7H1 G3ML32 A0A023GFU7 A0A1E1WZF1 A0A131XJP9 A0A2B4RZB2 A0A026WAD7 A0A2Y9QLB7 A0A2Y9QFV8 A0A0K8R8Z2 A0A3Q7TLT2 A0A2Y9HZA2 A0A2U3ZGP9 A0A3Q7U4M6 U6DW12 A0A3Q7NN36 A0A2Y9HZQ3 A0A091DUX7 F6V5H3 A0A384C7V0 A0A3Q7NNG3 A0A2Y9HZA6 A0A3Q7WY86 L5JV16 A0A384C6T0 A0A3Q7XUM0 G1TK72 A0A1S3IJZ8 H0XQF3 D2HZ82 B7PLK7 V9L127 A0A250XW42 A0A2U3XPB0 G1Q6N8 K9IWQ8 A0A2K5RMM0 Q3T107 A0A2K6ARW3 A0A2K6LR30 A0A384C6V1 G1RQA7 A0A2K5YTH6 A0A0D9R986 A0A2R9BW20 A0A096MWW9 A0A2K5MWH9 A0A2U3W0R9 A0A2K5I1Z0 A0A3Q7W102 G7P700 G3RS69 H2QS35 G7MU36 Q969Q5 I3MWQ8 E2R4M1 F6XZH4 A0A2Y9DBD6 A0A3Q7NWZ5 A0A2K5CR06 A0A2Y9HZ97 A0A2U3XP12 A0A2I2UYV4 L5LY47 A0A3Q7T1R8
PDB
2HEI
E-value=5.03565e-11,
Score=156
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Only about 20-25% is recovered in the particulate fraction. With evidence from 2 publications.
Cytosol Only about 20-25% is recovered in the particulate fraction. With evidence from 2 publications.
Membrane Only about 20-25% is recovered in the particulate fraction. With evidence from 2 publications.
Cytosol Only about 20-25% is recovered in the particulate fraction. With evidence from 2 publications.
Membrane Only about 20-25% is recovered in the particulate fraction. With evidence from 2 publications.
Length:
77
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00394
Exp number, first 60 AAs:
0.00384
Total prob of N-in:
0.32566
outside
1 - 77
Population Genetic Test Statistics
Pi
212.12786
Theta
184.939799
Tajima's D
0.462591
CLR
59.982732
CSRT
0.499675016249188
Interpretation
Uncertain
