Gene
KWMTBOMO06274 Validated by peptides from experiments
Annotation
PREDICTED:_probable_cyclin-dependent_serine/threonine-protein_kinase_DDB_G0292550_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.229
Sequence
CDS
ATGTACCTTCATAAAAAAAGTAACGTGACGTATGAGGGCAAAGGGATCCTGAGCTGTTTTGATTATTTCGCAAATGGAATAAACAGAAAATGCGGTAATTCTTATAACATATTCACGGATATCGCGAAATTGAACGGTCGAGGCAACCACGATCAAGGAATAAAAGTCATAATCGACGTTTCCAAAGGGGGTGGACATATTTTAGATTACTGGATTTCGGGACTGGGATCTACATTCGCAGATATGGTTATATCGGCGCACAGAATCGTCATATGCGCATTTGGAAGGAAACATAGAAGAAGTTTCTTTTTTGCAGACAAAGGAATCTGCCGGAGTGACTTCGGGAAGCATTACGGTTGGCGAAGCTATGATAATGGCTATGATGATATAGCTAATGTTATAAGCTATTATCGCCTCAATAATGCGCAAACTGACATTGAGAATATTTTCAAGAAGCACACCAAAGGCAAAGGGATTAAAGCGTTGACAGGGAGTCACCGGTTAATCGCTCAAGCAATATGGGATTTATCGAAACTATTCAGTCGTGCAAACAATGAGTGGTGCCATGATGACGTATATTACGTTTTCAAAAGAGTCTTAACTGTATTAGGAGCTCTGGCAAATGGTTATAGAACTGGACTTGGAAACGATGTTGTTGATGGGCTGATCGAGTATTGCGGGGAAGCCGTCAAACAATACGGGCAAAAGGGTAGATCTATCCCACTGTGGACGACGATAGCGGTGACGATAGTAACGATAATGCTAATGATGATAACGGAAATAACGACGAATATAACAGGGACGAAAATAGCAGTGGCAGCACTAAGAGAAAAAGTAGTAGCAGTGACGAAAATAGCAGTGGCAGTAATAAAAATAGCAGTGCCAACACTAATAAAAAAAGCATTAGCAGTGACGAAAACAGCAGTGGCAGCACTAAGAGAAAAAGTAGTAGCAGTGACGAAAATAGCAGTAGCGGTGACGAAAATAGCAGTAGCAGTGACGACAATAGCAGTGGCAGTAATGAAAATAGCAGTGGCAACACTAATAAAAGAAGCAGTAGCAGTGACAAAAATAGTAGTAGCAATAATGAAAATAGCAGTGGCAATAGTAGCAAAAAAAGCAATAGCAGTAGCGAAAATAGCAGTAACCACAGTAGCAAAAAAAGCAATAGCAATGACGAAAATAGCAGTAGTAGTAACGAAAATAGCAGTGGCAACACTAGCCGAACGAGCAGCAGTAGCGCGAAGAAACATAGAAAACATAGCAGTGAAAATAATGATAGCAATACGGACACCAATAATAACACGAACAATAATGATTCAAATCAATCAAATCGATCAAGTAACTCCAACAATAATCGAAATAATCAAGAGACTAATATCGTTATCAACAATAATTATGTTGTTTATAATAAAGAAGAGCTCACTCTCAGAGTTTGTACGTACATTCACGTCATGA
Protein
MYLHKKSNVTYEGKGILSCFDYFANGINRKCGNSYNIFTDIAKLNGRGNHDQGIKVIIDVSKGGGHILDYWISGLGSTFADMVISAHRIVICAFGRKHRRSFFFADKGICRSDFGKHYGWRSYDNGYDDIANVISYYRLNNAQTDIENIFKKHTKGKGIKALTGSHRLIAQAIWDLSKLFSRANNEWCHDDVYYVFKRVLTVLGALANGYRTGLGNDVVDGLIEYCGEAVKQYGQKGRSIPLWTTIAVTIVTIMLMMITEITTNITGTKIAVAALREKVVAVTKIAVAVIKIAVPTLIKKALAVTKTAVAALREKVVAVTKIAVAVTKIAVAVTTIAVAVMKIAVATLIKEAVAVTKIVVAIMKIAVAIVAKKAIAVAKIAVTTVAKKAIAMTKIAVVVTKIAVATLAERAAVARRNIENIAVKIMIAIRTPIITRTIMIQINQIDQVTPTIIEIIKRLISLSTIIMLFIIKKSSLSEFVRTFTS
Summary
Uniprot
ProteinModelPortal
PDB
5X2Q
E-value=0.0745,
Score=85
Ontologies
GO
Topology
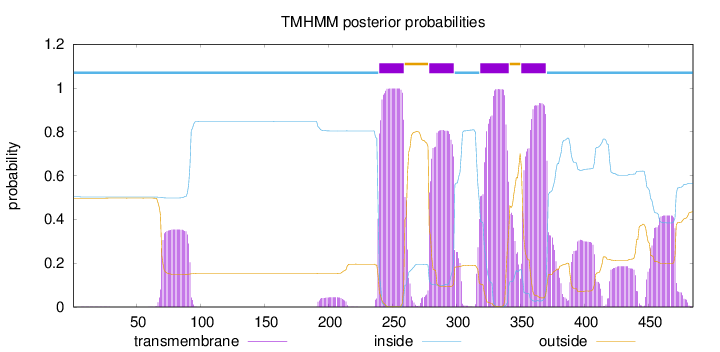
Length:
485
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
107.84699
Exp number, first 60 AAs:
0.02176
Total prob of N-in:
0.50311
inside
1 - 239
TMhelix
240 - 259
outside
260 - 278
TMhelix
279 - 298
inside
299 - 318
TMhelix
319 - 341
outside
342 - 350
TMhelix
351 - 370
inside
371 - 485
Population Genetic Test Statistics
Pi
178.836704
Theta
156.76295
Tajima's D
-0.537281
CLR
0.495304
CSRT
0.234788260586971
Interpretation
Uncertain
