Pre Gene Modal
BGIBMGA001818
Annotation
PREDICTED:_nuclear_pore_complex_protein_Nup98-Nup96_isoform_X1_[Bombyx_mori]
Full name
Nuclear pore complex protein Nup98-Nup96
Location in the cell
Cytoplasmic Reliability : 1.21 Nuclear Reliability : 1.371
Sequence
CDS
ATGAGACCGCCCCTGCCGGATTACAATGAAGATTACTGCTACATCACGAGTAAGAATGGAAATACGAAAGCGGTGTTGGATTTACGCTACGAGCTTATTAGAGCAAGAGCTTTCAATCAACGTCCGAAACTTAAGCCCGCAGCTTACACTCCGGATCCGAACGATTTTACACTGTGCTTCCTCCTGGGCGCGTGGTTCGGCAGTCCGTCGGTGGAGAGCGTGGTGTCGGTGGCGGAGCAGCTGGAGGCGGGCGGCGCGTGGCAGCTGGCCACCGCCGTGCTGGCGTACCATCCGCACGGAGCCGCCCGATCCTCGCTGGTGAGGAGCATGATCTCGCGGCACGCGCCGTCCTCGGGCTCCGCCCCCGGCCTCCAGCTGCTGGAGCGGATCGGGGTGCCCGAGAAGTGGATCCACCTGGCTCGCGCGCACAGGGCCAAGTACGAGCACCAGCCGGCGCTGGAGGTGGAGCATCTGTTGGCAGCGCGGCAGTGGAGCGCCGCGCACCGCGTGCTGCTGGACGAGCTGCTTCCCGACGCCGTGCTCAACGACGATCTGCAAACTGTATCCCGGCTGCTGAAGACGCTGGAGGAGGCGGCCGAGCGACACGAGGTGGCCGACTGGGAGAGCGGCGGGCTCGCGCTGTCCCACTACATACACGTCTGTGACGAGATAATGGCGATACAGAATGAAGGCGATCAGTGGGGACGGCTGGAGGCGCTTCGCCCCCGCCTGGCCGCCGCCTGTAGGGCGCTGCCCCCTCCACAACACATGTCGCCGCGTGTGGCCGCGGCCCGAGCGCAGATGGGGGGTCGCTTGCTGCGGCTGACGCTGTCCGCCAACCTGTCCGCCGACAAGCTGGCGTCGCTGGTGGCCGCCCTGAAGCTGCCCCCGGACTGCTCCGCCTGCGCCAGTCACAAGATAACAACAGAACTCGCCGAACGCAGATTAGAAGGGAATTATACGGCCCTGCTTGCACCTTAA
Protein
MRPPLPDYNEDYCYITSKNGNTKAVLDLRYELIRARAFNQRPKLKPAAYTPDPNDFTLCFLLGAWFGSPSVESVVSVAEQLEAGGAWQLATAVLAYHPHGAARSSLVRSMISRHAPSSGSAPGLQLLERIGVPEKWIHLARAHRAKYEHQPALEVEHLLAARQWSAAHRVLLDELLPDAVLNDDLQTVSRLLKTLEEAAERHEVADWESGGLALSHYIHVCDEIMAIQNEGDQWGRLEALRPRLAAACRALPPPQHMSPRVAAARAQMGGRLLRLTLSANLSADKLASLVAALKLPPDCSACASHKITTELAERRLEGNYTALLAP
Summary
Description
Plays a role in the nuclear pore complex (NPC) assembly and/or maintenance. NUP98 and NUP96 are involved in the bidirectional transport across the NPC. May anchor NUP153 and TPR to the NPC. In cooperation with DHX9, plays a role in transcription and alternative splicing activation of a subset of genes (PubMed:28221134). Involved in the localization of DHX9 in discrete intranuclear foci (GLFG-body) (PubMed:28221134).
Plays a role in the nuclear pore complex (NPC) assembly and/or maintenance. Involved in the bidirectional transport across the NPC. May anchor NUP153 and TPR to the NPC.
Plays a role in the nuclear pore complex (NPC) assembly and/or maintenance. NUP98 and NUP96 are involved in the bidirectional transport across the NPC. May anchor NUP153 and TPR to the NPC. In cooperation with DHX9, plays a role in transcription and alternative splicing activation of a subset of genes. Involved in the localization of DHX9 in discrete intranuclear foci (GLFG-body).
Plays a role in the nuclear pore complex (NPC) assembly and/or maintenance. Involved in the bidirectional transport across the NPC. May anchor NUP153 and TPR to the NPC.
Plays a role in the nuclear pore complex (NPC) assembly and/or maintenance. NUP98 and NUP96 are involved in the bidirectional transport across the NPC. May anchor NUP153 and TPR to the NPC. In cooperation with DHX9, plays a role in transcription and alternative splicing activation of a subset of genes. Involved in the localization of DHX9 in discrete intranuclear foci (GLFG-body).
Subunit
Part of the nuclear pore complex (NPC) (PubMed:15229283, PubMed:18287282). Interacts directly with NUP96 (PubMed:12191480). Part of the Nup160 subcomplex in the nuclear pore which is composed of NUP160, NUP133, NUP107 and NUP96; this complex plays a role in RNA export and in tethering NUP98 and NUP153 to the nucleus (PubMed:11684705). Interacts with RAE1 (PubMed:10209021, PubMed:20498086). Does not interact with TPR (PubMed:11684705). Interacts directly with NUP88 and NUP214, subunits of the cytoplasmic filaments of the NPC (By similarity). Interacts (via N-terminus) with DHX9 (via DRBM, OB-fold and RGG domains); this interaction occurs in a RNA-dependent manner and stimulates DHX9-mediated ATPase activity (PubMed:28221134).
(Microbial infection) Interacts with vesicular stomatitis virus protein M (PubMed:11106761).
Part of the nuclear pore complex (NPC). Interacts directly with NUP96. Part of the Nup160 subcomplex in the nuclear pore which is composed of NUP160, NUP133, NUP107 and NUP96; this complex plays a role in RNA export and in tethering NUP98 and NUP153 to the nucleus. Interacts with RAE1. Does not interact with TPR. Interacts directly with NUP88 and NUP214, subunits of the cytoplasmic filaments of the NPC. Interacts (via N-terminus) with DHX9 (via DRBM, OB-fold and RGG domains); this interaction occurs in a RNA-dependent manner and stimulates DHX9-mediated ATPase activity.
(Microbial infection) Interacts with vesicular stomatitis virus protein M (PubMed:11106761).
Part of the nuclear pore complex (NPC). Interacts directly with NUP96. Part of the Nup160 subcomplex in the nuclear pore which is composed of NUP160, NUP133, NUP107 and NUP96; this complex plays a role in RNA export and in tethering NUP98 and NUP153 to the nucleus. Interacts with RAE1. Does not interact with TPR. Interacts directly with NUP88 and NUP214, subunits of the cytoplasmic filaments of the NPC. Interacts (via N-terminus) with DHX9 (via DRBM, OB-fold and RGG domains); this interaction occurs in a RNA-dependent manner and stimulates DHX9-mediated ATPase activity.
Similarity
Belongs to the nucleoporin GLFG family.
Keywords
3D-structure
Acetylation
Alternative splicing
Autocatalytic cleavage
Chromosomal rearrangement
Complete proteome
Direct protein sequencing
Host-virus interaction
Hydrolase
Isopeptide bond
Membrane
mRNA transport
Nuclear pore complex
Nucleus
Phosphoprotein
Polymorphism
Protease
Protein transport
Reference proteome
Repeat
Serine protease
Translocation
Transport
Ubl conjugation
Feature
chain Nuclear pore complex protein Nup98
splice variant In isoform 2, isoform 4 and isoform 5.
sequence variant In a breast cancer sample; somatic mutation.
splice variant In isoform 2, isoform 4 and isoform 5.
sequence variant In a breast cancer sample; somatic mutation.
Uniprot
H9IX35
A0A194Q4W3
A0A2W1BLS0
A0A212F926
A0A2M4A068
A0A2M3ZZQ1
+ More
A0A182FR72 A0A1B0B8F3 A0A034V6D0 A0A1A9Y2K9 A0A336LFV7 T1EAL4 A0A2M3ZZY1 A0A091G603 A0A2L2Y5W2 A0A336KXZ5 A0A2M4B8W5 A0A093HLV1 A0A182VQ92 A0A1A9WVG8 T1IPI4 A0A182RCW1 A0A093BPU1 A0A0K8U4G4 A0A1A9VGF5 A0A1A9ZUP2 W5JD26 A0A0K8WGH0 A0A1B0FAH0 A0A0A1WUD9 A0A0A1WPK7 K9IPD0 K9J3Z7 A0A093IB35 A0A093F2W3 A0A2Y9SDG7 A0A2Y9MHS6 A0A2Y9SEW8 A0A2Y9MKI0 A0A2Y9MKI4 A0A2Y9MH87 A0A2Y9S6R3 A0A340XVD3 Q7QBX5 A0A341AEL1 A0A341AA59 A0A182XID2 G3SH93 A0A099Z778 U3K6I4 A0A182VF73 W5NVX9 W5NVY1 G1LWU9 E1BCV4 D2HYT4 A0A087VHJ3 L8J2T2 A0A182MCY3 A0A182K2G3 A0A1S3F062 A0A182Y7A1 A0A1S3F055 H0V7X8 A0A2M3ZFG0 A0A3Q7Y6Z4 P52948-6 A0A384DEQ7 A0A3Q7Y108 A0A3Q7WLC0 A0A3Q7Y6Z8 A0A384DER3 A0A384DF97 P52948 P52948-5 A0A3L8RTL3 A0A212DJ24 A0A218UH21 A0A091KE74 A0A087QNE8 A0A182PA24 H2NEI3 A0A2J8V286 I7G613 A0A0L0CEF9 L9LAX6 H9H2Z0 H7C3P6 P49793 A0A093T8F8 A0A2J8IU24 A0A3Q7TM51 A0A3Q7TYV9 F6R2Q1
A0A182FR72 A0A1B0B8F3 A0A034V6D0 A0A1A9Y2K9 A0A336LFV7 T1EAL4 A0A2M3ZZY1 A0A091G603 A0A2L2Y5W2 A0A336KXZ5 A0A2M4B8W5 A0A093HLV1 A0A182VQ92 A0A1A9WVG8 T1IPI4 A0A182RCW1 A0A093BPU1 A0A0K8U4G4 A0A1A9VGF5 A0A1A9ZUP2 W5JD26 A0A0K8WGH0 A0A1B0FAH0 A0A0A1WUD9 A0A0A1WPK7 K9IPD0 K9J3Z7 A0A093IB35 A0A093F2W3 A0A2Y9SDG7 A0A2Y9MHS6 A0A2Y9SEW8 A0A2Y9MKI0 A0A2Y9MKI4 A0A2Y9MH87 A0A2Y9S6R3 A0A340XVD3 Q7QBX5 A0A341AEL1 A0A341AA59 A0A182XID2 G3SH93 A0A099Z778 U3K6I4 A0A182VF73 W5NVX9 W5NVY1 G1LWU9 E1BCV4 D2HYT4 A0A087VHJ3 L8J2T2 A0A182MCY3 A0A182K2G3 A0A1S3F062 A0A182Y7A1 A0A1S3F055 H0V7X8 A0A2M3ZFG0 A0A3Q7Y6Z4 P52948-6 A0A384DEQ7 A0A3Q7Y108 A0A3Q7WLC0 A0A3Q7Y6Z8 A0A384DER3 A0A384DF97 P52948 P52948-5 A0A3L8RTL3 A0A212DJ24 A0A218UH21 A0A091KE74 A0A087QNE8 A0A182PA24 H2NEI3 A0A2J8V286 I7G613 A0A0L0CEF9 L9LAX6 H9H2Z0 H7C3P6 P49793 A0A093T8F8 A0A2J8IU24 A0A3Q7TM51 A0A3Q7TYV9 F6R2Q1
EC Number
3.4.21.-
Pubmed
19121390
26354079
28756777
22118469
25348373
26561354
+ More
20920257 23761445 25830018 12364791 14747013 17210077 22398555 20809919 20010809 19393038 22751099 25244985 21993624 8563754 10087256 16554811 15489334 17974005 10556215 10209021 11106761 11493482 11684705 11986249 11839768 12802065 15229283 16028218 15725483 17081983 16467868 16964243 17287853 18220336 18691976 18669648 18318008 19413330 19690332 19608861 20407419 20068231 21269460 21406692 22058212 23186163 24275569 25944712 28221134 28112733 12191480 18287282 20498086 16959974 24813606 30282656 29294181 17194215 26108605 23385571 17431167 7736573 22673903 18464734
20920257 23761445 25830018 12364791 14747013 17210077 22398555 20809919 20010809 19393038 22751099 25244985 21993624 8563754 10087256 16554811 15489334 17974005 10556215 10209021 11106761 11493482 11684705 11986249 11839768 12802065 15229283 16028218 15725483 17081983 16467868 16964243 17287853 18220336 18691976 18669648 18318008 19413330 19690332 19608861 20407419 20068231 21269460 21406692 22058212 23186163 24275569 25944712 28221134 28112733 12191480 18287282 20498086 16959974 24813606 30282656 29294181 17194215 26108605 23385571 17431167 7736573 22673903 18464734
EMBL
BABH01021286
KQ459463
KPJ00394.1
KZ150084
PZC73806.1
AGBW02009652
+ More
OWR50231.1 GGFK01000717 MBW34038.1 GGFK01000706 MBW34027.1 JXJN01009920 GAKP01021834 JAC37118.1 UFQS01003267 UFQT01003267 SSX15435.1 SSX34804.1 GAMD01000933 JAB00658.1 GGFK01000744 MBW34065.1 KL447080 KFO69642.1 IAAA01009455 IAAA01009456 IAAA01009457 LAA02700.1 UFQS01001329 UFQT01001329 SSX10382.1 SSX30070.1 GGFJ01000127 MBW49268.1 KL206203 KFV79957.1 JH431263 KL451229 KFV05760.1 GDHF01030923 JAI21391.1 ADMH02001587 ETN61956.1 GDHF01002112 JAI50202.1 CCAG010015281 GBXI01012157 JAD02135.1 GBXI01013969 JAD00323.1 GABZ01003500 JAA50025.1 GABZ01003506 JAA50019.1 KK590751 KFV99924.1 KK620339 KFV50835.1 AAAB01008859 EAA08227.5 CABD030077206 CABD030077207 CABD030077208 CABD030077209 KL891034 KGL78314.1 AGTO01012089 AMGL01032342 ACTA01002145 ACTA01010145 ACTA01018145 ACTA01026145 ACTA01034145 ACTA01042144 ACTA01050144 ACTA01178136 ACTA01186134 ACTA01194132 GL193731 EFB29397.1 KL493705 KFO12085.1 JH880247 ELR63046.1 AXCM01001786 AAKN02051231 AAKN02051232 AAKN02051233 GGFM01006545 MBW27296.1 U41815 AB040538 AF071076 AF071077 AF231130 AC060812 AC090587 BC041136 BC012906 AF116074 BT007349 AL133601 AL137613 QUSF01000341 RLV82888.1 MKHE01000001 OWK18154.1 MUZQ01000308 OWK53034.1 KK502284 KFP22051.1 KL225752 KFM02752.1 ABGA01156939 ABGA01156940 ABGA01156941 ABGA01156942 ABGA01156943 ABGA01156944 ABGA01156945 ABGA01156946 ABGA01156947 ABGA01156948 NDHI03003437 PNJ51639.1 AB172654 BAE89716.1 JRES01000501 KNC30640.1 KB320452 ELW71834.1 JSUE03012860 JSUE03012861 JSUE03012862 JSUE03012863 JSUE03012864 JSUE03012865 JSUE03012866 JSUE03012867 JSUE03012868 CH473956 L39991 KL428082 KFW90844.1 NBAG03000591 PNI13999.1
OWR50231.1 GGFK01000717 MBW34038.1 GGFK01000706 MBW34027.1 JXJN01009920 GAKP01021834 JAC37118.1 UFQS01003267 UFQT01003267 SSX15435.1 SSX34804.1 GAMD01000933 JAB00658.1 GGFK01000744 MBW34065.1 KL447080 KFO69642.1 IAAA01009455 IAAA01009456 IAAA01009457 LAA02700.1 UFQS01001329 UFQT01001329 SSX10382.1 SSX30070.1 GGFJ01000127 MBW49268.1 KL206203 KFV79957.1 JH431263 KL451229 KFV05760.1 GDHF01030923 JAI21391.1 ADMH02001587 ETN61956.1 GDHF01002112 JAI50202.1 CCAG010015281 GBXI01012157 JAD02135.1 GBXI01013969 JAD00323.1 GABZ01003500 JAA50025.1 GABZ01003506 JAA50019.1 KK590751 KFV99924.1 KK620339 KFV50835.1 AAAB01008859 EAA08227.5 CABD030077206 CABD030077207 CABD030077208 CABD030077209 KL891034 KGL78314.1 AGTO01012089 AMGL01032342 ACTA01002145 ACTA01010145 ACTA01018145 ACTA01026145 ACTA01034145 ACTA01042144 ACTA01050144 ACTA01178136 ACTA01186134 ACTA01194132 GL193731 EFB29397.1 KL493705 KFO12085.1 JH880247 ELR63046.1 AXCM01001786 AAKN02051231 AAKN02051232 AAKN02051233 GGFM01006545 MBW27296.1 U41815 AB040538 AF071076 AF071077 AF231130 AC060812 AC090587 BC041136 BC012906 AF116074 BT007349 AL133601 AL137613 QUSF01000341 RLV82888.1 MKHE01000001 OWK18154.1 MUZQ01000308 OWK53034.1 KK502284 KFP22051.1 KL225752 KFM02752.1 ABGA01156939 ABGA01156940 ABGA01156941 ABGA01156942 ABGA01156943 ABGA01156944 ABGA01156945 ABGA01156946 ABGA01156947 ABGA01156948 NDHI03003437 PNJ51639.1 AB172654 BAE89716.1 JRES01000501 KNC30640.1 KB320452 ELW71834.1 JSUE03012860 JSUE03012861 JSUE03012862 JSUE03012863 JSUE03012864 JSUE03012865 JSUE03012866 JSUE03012867 JSUE03012868 CH473956 L39991 KL428082 KFW90844.1 NBAG03000591 PNI13999.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000069272
UP000092460
UP000092443
+ More
UP000053760 UP000053584 UP000075920 UP000091820 UP000075900 UP000078200 UP000092445 UP000000673 UP000092444 UP000248483 UP000265300 UP000007062 UP000252040 UP000076407 UP000001519 UP000053641 UP000016665 UP000075903 UP000002356 UP000008912 UP000009136 UP000075883 UP000075881 UP000081671 UP000076408 UP000005447 UP000286642 UP000005640 UP000261680 UP000291021 UP000276834 UP000197619 UP000053119 UP000053286 UP000075885 UP000001595 UP000037069 UP000011518 UP000006718 UP000002494 UP000286640 UP000002279
UP000053760 UP000053584 UP000075920 UP000091820 UP000075900 UP000078200 UP000092445 UP000000673 UP000092444 UP000248483 UP000265300 UP000007062 UP000252040 UP000076407 UP000001519 UP000053641 UP000016665 UP000075903 UP000002356 UP000008912 UP000009136 UP000075883 UP000075881 UP000081671 UP000076408 UP000005447 UP000286642 UP000005640 UP000261680 UP000291021 UP000276834 UP000197619 UP000053119 UP000053286 UP000075885 UP000001595 UP000037069 UP000011518 UP000006718 UP000002494 UP000286640 UP000002279
Interpro
SUPFAM
SSF82215
SSF82215
Gene 3D
ProteinModelPortal
H9IX35
A0A194Q4W3
A0A2W1BLS0
A0A212F926
A0A2M4A068
A0A2M3ZZQ1
+ More
A0A182FR72 A0A1B0B8F3 A0A034V6D0 A0A1A9Y2K9 A0A336LFV7 T1EAL4 A0A2M3ZZY1 A0A091G603 A0A2L2Y5W2 A0A336KXZ5 A0A2M4B8W5 A0A093HLV1 A0A182VQ92 A0A1A9WVG8 T1IPI4 A0A182RCW1 A0A093BPU1 A0A0K8U4G4 A0A1A9VGF5 A0A1A9ZUP2 W5JD26 A0A0K8WGH0 A0A1B0FAH0 A0A0A1WUD9 A0A0A1WPK7 K9IPD0 K9J3Z7 A0A093IB35 A0A093F2W3 A0A2Y9SDG7 A0A2Y9MHS6 A0A2Y9SEW8 A0A2Y9MKI0 A0A2Y9MKI4 A0A2Y9MH87 A0A2Y9S6R3 A0A340XVD3 Q7QBX5 A0A341AEL1 A0A341AA59 A0A182XID2 G3SH93 A0A099Z778 U3K6I4 A0A182VF73 W5NVX9 W5NVY1 G1LWU9 E1BCV4 D2HYT4 A0A087VHJ3 L8J2T2 A0A182MCY3 A0A182K2G3 A0A1S3F062 A0A182Y7A1 A0A1S3F055 H0V7X8 A0A2M3ZFG0 A0A3Q7Y6Z4 P52948-6 A0A384DEQ7 A0A3Q7Y108 A0A3Q7WLC0 A0A3Q7Y6Z8 A0A384DER3 A0A384DF97 P52948 P52948-5 A0A3L8RTL3 A0A212DJ24 A0A218UH21 A0A091KE74 A0A087QNE8 A0A182PA24 H2NEI3 A0A2J8V286 I7G613 A0A0L0CEF9 L9LAX6 H9H2Z0 H7C3P6 P49793 A0A093T8F8 A0A2J8IU24 A0A3Q7TM51 A0A3Q7TYV9 F6R2Q1
A0A182FR72 A0A1B0B8F3 A0A034V6D0 A0A1A9Y2K9 A0A336LFV7 T1EAL4 A0A2M3ZZY1 A0A091G603 A0A2L2Y5W2 A0A336KXZ5 A0A2M4B8W5 A0A093HLV1 A0A182VQ92 A0A1A9WVG8 T1IPI4 A0A182RCW1 A0A093BPU1 A0A0K8U4G4 A0A1A9VGF5 A0A1A9ZUP2 W5JD26 A0A0K8WGH0 A0A1B0FAH0 A0A0A1WUD9 A0A0A1WPK7 K9IPD0 K9J3Z7 A0A093IB35 A0A093F2W3 A0A2Y9SDG7 A0A2Y9MHS6 A0A2Y9SEW8 A0A2Y9MKI0 A0A2Y9MKI4 A0A2Y9MH87 A0A2Y9S6R3 A0A340XVD3 Q7QBX5 A0A341AEL1 A0A341AA59 A0A182XID2 G3SH93 A0A099Z778 U3K6I4 A0A182VF73 W5NVX9 W5NVY1 G1LWU9 E1BCV4 D2HYT4 A0A087VHJ3 L8J2T2 A0A182MCY3 A0A182K2G3 A0A1S3F062 A0A182Y7A1 A0A1S3F055 H0V7X8 A0A2M3ZFG0 A0A3Q7Y6Z4 P52948-6 A0A384DEQ7 A0A3Q7Y108 A0A3Q7WLC0 A0A3Q7Y6Z8 A0A384DER3 A0A384DF97 P52948 P52948-5 A0A3L8RTL3 A0A212DJ24 A0A218UH21 A0A091KE74 A0A087QNE8 A0A182PA24 H2NEI3 A0A2J8V286 I7G613 A0A0L0CEF9 L9LAX6 H9H2Z0 H7C3P6 P49793 A0A093T8F8 A0A2J8IU24 A0A3Q7TM51 A0A3Q7TYV9 F6R2Q1
PDB
5A9Q
E-value=3.0529e-18,
Score=224
Ontologies
GO
GO:0005643
GO:0017056
GO:0031080
GO:0016973
GO:0044614
GO:0006405
GO:0006606
GO:0034398
GO:0003723
GO:0008139
GO:0000973
GO:0003729
GO:1990904
GO:0034399
GO:0000776
GO:0003713
GO:0042405
GO:1990841
GO:0048026
GO:0006260
GO:0051292
GO:0016604
GO:0044615
GO:0031965
GO:0060964
GO:0008236
GO:0005635
GO:0019083
GO:0006409
GO:0005215
GO:0005829
GO:0005654
GO:0006406
GO:0043657
GO:0043231
GO:0006110
GO:0075733
GO:0006999
GO:0016925
GO:0016032
GO:1900034
GO:0006913
GO:0051028
GO:0042277
PANTHER
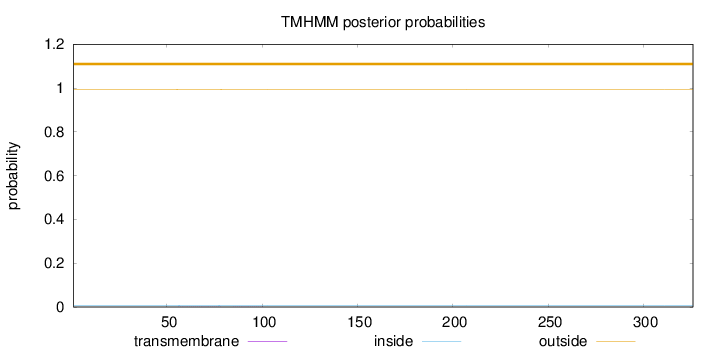
Topology
Subcellular location
Nucleus membrane
Localized to the nucleoplasmic side of the nuclear pore complex (NPC), at or near the nucleoplasmic basket (PubMed:11839768). Dissociates from the dissasembled NPC structure early during prophase of mitosis (PubMed:12802065). Colocalized with NUP153 and TPR to the nuclear basket of NPC (PubMed:11839768). Colocalized with DHX9 in diffuse and discrete intranuclear foci (GLFG-body) (PubMed:11839768, PubMed:28221134). Remains localized to the nuclear membrane after poliovirus (PV) infection (PubMed:11106761). With evidence from 21 publications.
Nucleus Localized to the nucleoplasmic side of the nuclear pore complex (NPC), at or near the nucleoplasmic basket (PubMed:11839768). Dissociates from the dissasembled NPC structure early during prophase of mitosis (PubMed:12802065). Colocalized with NUP153 and TPR to the nuclear basket of NPC (PubMed:11839768). Colocalized with DHX9 in diffuse and discrete intranuclear foci (GLFG-body) (PubMed:11839768, PubMed:28221134). Remains localized to the nuclear membrane after poliovirus (PV) infection (PubMed:11106761). With evidence from 21 publications.
Nuclear pore complex Localized to the nucleoplasmic side of the nuclear pore complex (NPC), at or near the nucleoplasmic basket (PubMed:11839768). Dissociates from the dissasembled NPC structure early during prophase of mitosis (PubMed:12802065). Colocalized with NUP153 and TPR to the nuclear basket of NPC (PubMed:11839768). Colocalized with DHX9 in diffuse and discrete intranuclear foci (GLFG-body) (PubMed:11839768, PubMed:28221134). Remains localized to the nuclear membrane after poliovirus (PV) infection (PubMed:11106761). With evidence from 21 publications.
Nucleoplasm Localized to the nucleoplasmic side of the nuclear pore complex (NPC), at or near the nucleoplasmic basket (PubMed:11839768). Dissociates from the dissasembled NPC structure early during prophase of mitosis (PubMed:12802065). Colocalized with NUP153 and TPR to the nuclear basket of NPC (PubMed:11839768). Colocalized with DHX9 in diffuse and discrete intranuclear foci (GLFG-body) (PubMed:11839768, PubMed:28221134). Remains localized to the nuclear membrane after poliovirus (PV) infection (PubMed:11106761). With evidence from 21 publications.
Nucleus Localized to the nucleoplasmic side of the nuclear pore complex (NPC), at or near the nucleoplasmic basket (PubMed:11839768). Dissociates from the dissasembled NPC structure early during prophase of mitosis (PubMed:12802065). Colocalized with NUP153 and TPR to the nuclear basket of NPC (PubMed:11839768). Colocalized with DHX9 in diffuse and discrete intranuclear foci (GLFG-body) (PubMed:11839768, PubMed:28221134). Remains localized to the nuclear membrane after poliovirus (PV) infection (PubMed:11106761). With evidence from 21 publications.
Nuclear pore complex Localized to the nucleoplasmic side of the nuclear pore complex (NPC), at or near the nucleoplasmic basket (PubMed:11839768). Dissociates from the dissasembled NPC structure early during prophase of mitosis (PubMed:12802065). Colocalized with NUP153 and TPR to the nuclear basket of NPC (PubMed:11839768). Colocalized with DHX9 in diffuse and discrete intranuclear foci (GLFG-body) (PubMed:11839768, PubMed:28221134). Remains localized to the nuclear membrane after poliovirus (PV) infection (PubMed:11106761). With evidence from 21 publications.
Nucleoplasm Localized to the nucleoplasmic side of the nuclear pore complex (NPC), at or near the nucleoplasmic basket (PubMed:11839768). Dissociates from the dissasembled NPC structure early during prophase of mitosis (PubMed:12802065). Colocalized with NUP153 and TPR to the nuclear basket of NPC (PubMed:11839768). Colocalized with DHX9 in diffuse and discrete intranuclear foci (GLFG-body) (PubMed:11839768, PubMed:28221134). Remains localized to the nuclear membrane after poliovirus (PV) infection (PubMed:11106761). With evidence from 21 publications.
Length:
326
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01182
Exp number, first 60 AAs:
0.002
Total prob of N-in:
0.00734
outside
1 - 326
Population Genetic Test Statistics
Pi
177.461745
Theta
143.315462
Tajima's D
0.079136
CLR
0.173129
CSRT
0.390430478476076
Interpretation
Uncertain
