Gene
KWMTBOMO06266
Annotation
PREDICTED:_nuclear_pore_complex_protein_Nup98-Nup96_isoform_X1_[Bombyx_mori]
Full name
Nuclear pore complex protein Nup98-Nup96
Location in the cell
Nuclear Reliability : 3.196
Sequence
CDS
ATGATAAACTACATGCGGCCGGACGGCACGTGTCCTGTTCCGCACCTGACGATCGGACGCAAGAACTACGGGAACGTGTACTACGACTCGGAGATCGACGTGGCCGGCCTCGACCTGGACGCCCTGGTGCACTTCCTGAACAAGGAGGTGATAGTCTATCCGGAGGACAGCGACAAGCCGCCGGTCGGAGTCGGCCTGAACAGGCGGGCCATCGTGACCCTCGACCGGGTGTGGCCCCGGGACAAGACCGAGAAACGACTCATCACCGAGCCCGACAGATTACTAAAACTGGATTACGAAGGGAAGCTGCGCAGGGTTTGCGAGAAACATGACACCAAGTTTATCGAGTATCGACCTCAAACGGGCAGCTGGGTGTTCAGGGTGGAGCATTTCAGTAAGTACGGTCTCACTGATTCCGACGAGGAGGACGACCCCACTCCGGACGTCCTGAAGCGGCAACTCCTCACCGAGTCTCTACAGAAAGCCAGCGCACCAGGTCCCAAGTCGCCTGTGAATGTGTCGGAGGCGGGGGCTGCGGTCCCGGGGCTCTTGGGGCCCGGACATCCCGGGCTGGGGGGCTCCGGCCCGCCAGGCCTGCTCAGCGAGGACAGCCTCTACAATCTCCACCACACTTCCTTACTGACTCTAAACGGCACCAGCAAAGGTGAGACAATATGA
Protein
MINYMRPDGTCPVPHLTIGRKNYGNVYYDSEIDVAGLDLDALVHFLNKEVIVYPEDSDKPPVGVGLNRRAIVTLDRVWPRDKTEKRLITEPDRLLKLDYEGKLRRVCEKHDTKFIEYRPQTGSWVFRVEHFSKYGLTDSDEEDDPTPDVLKRQLLTESLQKASAPGPKSPVNVSEAGAAVPGLLGPGHPGLGGSGPPGLLSEDSLYNLHHTSLLTLNGTSKGETI
Summary
Description
Part of the nuclear pore complex (NPC) (PubMed:25197089). Required for MAD import as part of the Nup107-160 complex and required for nuclear export of Moe probably via its association with Rae1 (PubMed:20547758, PubMed:28554770). Plays a role in nuclear mRNA export (PubMed:28554770). Promotes cell antiviral response by upregulating FoxK-dependent antiviral gene transcription (PubMed:25197089, PubMed:25852164). In germline stem cells, involved in their maintenance and division together with the TGF-Beta and EGFR signaling pathways (PubMed:21949861). In larval lymph glands, has a role in the maintenance of hematopoiesis by regulating Pvr expression (PubMed:25201876).
Nuclear pore complex protein Nup98: Part of the nuclear pore complex (NPC) (PubMed:25310983). In the nucleoplasm, binds to transcriptionally active chromatin with a preference for regulatory regions; co-localizes with RNA polymerase II in a RNA-independent manner and before transition into transcription elongation (PubMed:20144760, PubMed:20144761, PubMed:28366641). Plays a role in the transcriptional memory process by stabilizing enhancer-promoter loops and by mediating anchoring of chromatin to the nuclear pore complex region (PubMed:28366641). During larval development, interacts with trx and MBD-R2 and regulates transcription of developmental genes including ecdysone-responsive genes such as Eip74 and E23 (PubMed:20144761, PubMed:25310983, PubMed:28366641).
Nuclear pore complex protein Nup96: Part of the nuclear pore complex (NPC).
Nuclear pore complex protein Nup98: Part of the nuclear pore complex (NPC) (PubMed:25310983). In the nucleoplasm, binds to transcriptionally active chromatin with a preference for regulatory regions; co-localizes with RNA polymerase II in a RNA-independent manner and before transition into transcription elongation (PubMed:20144760, PubMed:20144761, PubMed:28366641). Plays a role in the transcriptional memory process by stabilizing enhancer-promoter loops and by mediating anchoring of chromatin to the nuclear pore complex region (PubMed:28366641). During larval development, interacts with trx and MBD-R2 and regulates transcription of developmental genes including ecdysone-responsive genes such as Eip74 and E23 (PubMed:20144761, PubMed:25310983, PubMed:28366641).
Nuclear pore complex protein Nup96: Part of the nuclear pore complex (NPC).
Subunit
Part of the nuclear pore complex (NPC) (PubMed:25310983). Interacts with Rae1 (PubMed:21874015, PubMed:28554770). Nuclear pore complex protein Nup98: Interacts with pzg and Chro (PubMed:25310983). Interacts with MBD-R2; the interaction allows Nup98 recruitment to chromatin (PubMed:25310983). Interacts with Trx (PubMed:25310983). Interacts with Wds (PubMed:25310983). Interacts with Mtor and Cp190 (PubMed:28366641). Upon ecdysone stimulation, interacts with EcR, CTCF, su(Hw) and Trl (PubMed:28366641).
Similarity
Belongs to the nucleoporin GLFG family.
Keywords
Alternative splicing
Autocatalytic cleavage
Chromosome
Complete proteome
Hydrolase
Membrane
mRNA transport
Nuclear pore complex
Nucleus
Protease
Protein transport
Reference proteome
Repeat
Serine protease
Transcription
Transcription regulation
Translocation
Transport
Feature
chain Nuclear pore complex protein Nup98
splice variant In isoform C.
splice variant In isoform C.
Uniprot
EC Number
3.4.21.-
Pubmed
EMBL
KZ150084
PZC73810.1
NWSH01000160
PCG78936.1
AGBW02008966
OWR52038.1
+ More
KQ459463 KPJ00394.1 AGBW02009652 OWR50231.1 BABH01021286 AY250799 AAP13294.1 CM000364 EDX14109.1 CM000160 EDW98192.1 AE014297 BT056313 BT011076 AAR82742.1 ACL68760.1 CH902617 EDV43949.1 CH477302 EAT44225.1 CH477450 EAT40699.1 GFDL01012515 JAV22530.1 GFDL01012591 JAV22454.1 GFDL01012538 JAV22507.1
KQ459463 KPJ00394.1 AGBW02009652 OWR50231.1 BABH01021286 AY250799 AAP13294.1 CM000364 EDX14109.1 CM000160 EDW98192.1 AE014297 BT056313 BT011076 AAR82742.1 ACL68760.1 CH902617 EDV43949.1 CH477302 EAT44225.1 CH477450 EAT40699.1 GFDL01012515 JAV22530.1 GFDL01012591 JAV22454.1 GFDL01012538 JAV22507.1
Proteomes
Interpro
SUPFAM
SSF82215
SSF82215
Gene 3D
ProteinModelPortal
PDB
2Q5X
E-value=2.1611e-39,
Score=405
Ontologies
GO
GO:0017056
GO:0005643
GO:0071733
GO:0043922
GO:0000785
GO:0005737
GO:0035080
GO:1990841
GO:0006405
GO:0006606
GO:0005704
GO:0030838
GO:0035167
GO:0071390
GO:0060261
GO:0005703
GO:0036098
GO:0031490
GO:0002230
GO:0008236
GO:0010628
GO:0031965
GO:0044613
GO:0005634
GO:0005654
GO:0051028
GO:0035075
GO:0005700
GO:0045944
GO:0034605
GO:0030718
GO:0006810
GO:0006378
GO:0016787
GO:0008235
GO:0019538
GO:0030145
GO:0015923
GO:0006260
GO:0006913
PANTHER
Topology
Subcellular location
Chromosome
Associates with transcriptionally active chromatin. With evidence from 4 publications.
Nucleus Associates with transcriptionally active chromatin. With evidence from 4 publications.
Nucleoplasm Associates with transcriptionally active chromatin. With evidence from 4 publications.
Nucleus membrane Associates with transcriptionally active chromatin. With evidence from 4 publications.
Nuclear pore complex Associates with transcriptionally active chromatin. With evidence from 4 publications.
Nucleus Associates with transcriptionally active chromatin. With evidence from 4 publications.
Nucleoplasm Associates with transcriptionally active chromatin. With evidence from 4 publications.
Nucleus membrane Associates with transcriptionally active chromatin. With evidence from 4 publications.
Nuclear pore complex Associates with transcriptionally active chromatin. With evidence from 4 publications.
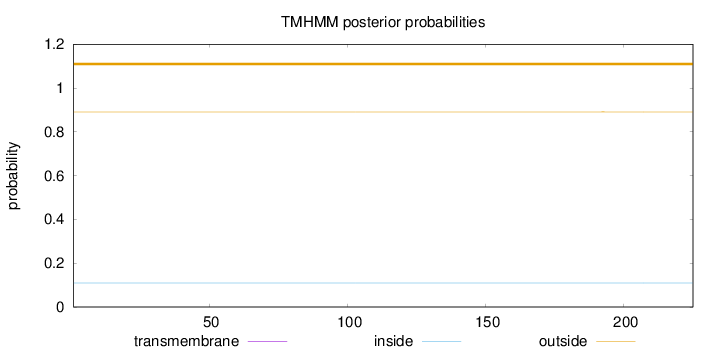
Length:
225
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00111
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10909
outside
1 - 225
Population Genetic Test Statistics
Pi
188.733706
Theta
147.324851
Tajima's D
1.311991
CLR
0.157804
CSRT
0.747212639368032
Interpretation
Uncertain
