Gene
KWMTBOMO06246
Pre Gene Modal
BGIBMGA001808
Annotation
PREDICTED:_uncharacterized_protein_LOC106133708_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.142
Sequence
CDS
ATGGCGTCTCAGATGCGTTCGCTCGCCGAGGAGGGGAAGGCGGGCGACGGTGCGGACGAGATGCTCCTGGATAAGAGCAGGGTGCTGGTCGTGCCGGCGGGGAAGAGTGGGGACGAGCACGCCCTGGTGACGGTTAAATCTTCCAAAAAAGTAAAGCTGCCGGATGACGACACGCCGGCCGTCGGAGCGGGTCGATTCACGATCGGTCCGGCGCCGGAACGCGACTGGGAGATGGTCCCGCCCGATGGAAAGTGGGGCTGGTTCATCCTTGTCGGTGCGACCCTGGTCAATATACTGATACCGGGTACGATAAAGTCCTTCGGTGTGCTGATGGTGGAGTTTAATGATGTCTTCCACTCGAGTGCTTCTGCCTCGTCAGGGATCGTGGCCCTCTGCTATTTCCTGTACAGCAGCTTAGGTCCACTTTCGTCAATTCTATCCGTGAGGTGGTCATACAGGACTGTCACACTGATCGGGGGTAGTTTCGCAGCGTTTGGAATGATACTGAGTAGCGCAGCATTTTCTATAACCTATTTGTATTTTAGCTTCGGTGCTATGGTAGGTACGGGCGCTGGCCTCGCCTTCCCAGCCACAGTGTACATAGTCACTTCATACTTCGTGAGGTTCCGTGGTCTGGCGAACGGGATCTGCATGTCGGGCTCCGCGTTCGGCAGCATCATCCTGCCCCCGGTGCTTAGGTACCTGCTCGAGACCTACGGCTACAAAGGCGCCGTCTTGATATTGGGTGGAATAATGCTGAACGTTTGGGCGGCTGCTCTCCTGTTCCAGCCTGTCGAAGAGCACATGGTCAGGAAGTACAAGGAAAACCAGGAGGAGGACTGTCCTCAGGACGCCCTGCTAGAGGAACAAGAAGAGGAACAAGAGATTGACGTAATCGAAATGTCGCAGACGAACGATAAAAAGGGCATATCGGAAAACGGCAGGGCAGAGGAGAGTGGACTCGTCCACCAGGACAGCGTGCCCAGCATCGCGCCGTCCCAATCAAATAACCAACTGAAGCCGAATATGTCCAAACGGAAGTTGTCGTACTCGAAAACGCCCAAAGCGAATCCGAGCACGTCCTCGTTCAGCAGACACGGCTCGCAGGAAACCTTCAGCAGGAGGCAGAGCAACGCCGGACCGATCAGGAATTATTCGACGTCCAGCTTAGCTTACATATCCACCCCTTTCCACGGTTCGACTCTGTCAGCCTTTGAGACCCCCAACGAGTTCGCGTCCCAGTTCTCGTTGAAGTCCGTGGCGGAGAGCGTTACCGAGGCCGCCTACTGTTGCCTGTGCTGCAAGAAGAAAAAAAAGAAAAGCGAAAAGGAATCGTCGAAGTTCTTCGATCTGAGCCTGCTGACCGATCCCGCGTATCTTGTGGTGCTGCTATCGAGCGGCGCCGTCGCGATATCGGTGACGATCTTCATCATCAACCTGCCGTCGCACGCGGAGAACTTGGGGTTCAGTAAGGCCAGGGGAGCTTACCTGCTCAGTACCGTGTCCACCCTCGACCTGGTCGGTCGCATCGGCGGCTCGACGCTCTGCGACCTGAACCTGATCCCGAAGTGGTGCTACTTCGTTGGCGGTCTGATATTCGCCGGCATATCTCTGATCATACTCCCGTTCATGGTTTCGTATACGTGGATAAGCGTCGTGTGCGCGTTCTACGGACTCGCCTCCGGAGTCAACAACAGCGTGACGACCCTCGTCCTTGCCGAGATATTGGGTGTCGAGCGGCTGATGTCCAGTTTCGGTATAAGCCTGTTCGTCAACGGCATCCTTCAGCTGATAGGGCCACCCCTCGGCGGTCTCTGGTTCGAGAGGGACCAGAACTATTTCCACATGTTCATAGCGCTCGGCTTTATATTCCTATTCGGGGCCTCCCTCTGGCTGTTCATGCCTTTCATAAATAGGCACAAGAATAAGAAGAAGTTGATGAATGATAAACTAACAACAGGCAAACTGGAGCCTTTCGCCGAGGAGGAAGCTGAGGAGGTGCAGGACGAACGGAATCCGAAGGTCGTCTTCACGCCCGAGGAGACCCCGTCCCAGATGAACGGCTCCCCGCCGATGTTCAAGAACAAATCGAACTCCAACTTCGAGGACGCGACTTCGCCGAGCAAGGACTTCGCTCGTTCGGCCAGCGCGATCCAGGTCGCCCAGAACAACGAGCGCTTCCGAAAGGTCTCGACGCCAGCCTCCTCAAGGCACGGCCTGCAAAACATCTCCAGCCGGAGCCACTTGCCCTCGACCCCATCCCTGCTTGGATCCGTCCCGGAAGGCAAGGCCAGTCGCGTTAACTCGCAAGAGGCGTTCGGAAAGAAGCTAGCCATCCCCAAGACCCCTAAACGCAGCCCTTCCACGTCCAGTTTCCAGTACATGTCGACCCCCTTCCACGGATCAACGCTGTCCGCCTTCGAGAAGCCCAGCGAATTCGCTTCCCAGTTCAGCCTCAAGTCGGTAACGGACAGTCTCGCCCCGATCTCCTACTGCTGCGGCTGTAAAAAGAAAGAGGACGACCCCAAAAAGGAGCCGAATCGTTATTTCGACGTCCAGCTACTGAAGGACCCCATTTATCTGGTCATACTTATATCTAATTGCACCTGTGCTATATCGTACACGAACTTCATAATATTAGTGCCGGCTTACGCCAATGAGTGCGGCTTCGACAAGTCTCTGGGCGCGTATTTGCTGTCGATCATATCCGCGTTGGATCTGGTCGGTCGGATCGGAGGCTCGGCTCTGTCCGACGTGGTTTCGACGCCGAAGCGTTACTTTTTCATATTCGGTTTGCTGTTGTCCGGCGTCAGCTTGTCTCTGATACCGTTGGTGCGGAGCTATTCGGCTATTTCCGCGTTCTGCTCGATTTTCGGCATCGCTTCGGGCATCAACGTCGGCGTCACGGCCCTGGTCATGACCGAAATGCTGGGTACAGAGCGACTAATGTCCTCGTACGGGATCTCGTTGTTCATGAACGGGATCTTGCAGCTGATCGGGCCGCCGATCTGCGGCTTCTGGTTCGAATACGCGAAGTCATATAAGTCCTTGTTCCTGACCTTCGGGCTGATCCTGATCGGAGGAGCTGCCCTGTGGGGACTCGTGCCCATCATCAATCGAAGGAGACGGCTGGCCGAACAAAACAAGCAAACCGTCGGGAAAGCTTAA
Protein
MASQMRSLAEEGKAGDGADEMLLDKSRVLVVPAGKSGDEHALVTVKSSKKVKLPDDDTPAVGAGRFTIGPAPERDWEMVPPDGKWGWFILVGATLVNILIPGTIKSFGVLMVEFNDVFHSSASASSGIVALCYFLYSSLGPLSSILSVRWSYRTVTLIGGSFAAFGMILSSAAFSITYLYFSFGAMVGTGAGLAFPATVYIVTSYFVRFRGLANGICMSGSAFGSIILPPVLRYLLETYGYKGAVLILGGIMLNVWAAALLFQPVEEHMVRKYKENQEEDCPQDALLEEQEEEQEIDVIEMSQTNDKKGISENGRAEESGLVHQDSVPSIAPSQSNNQLKPNMSKRKLSYSKTPKANPSTSSFSRHGSQETFSRRQSNAGPIRNYSTSSLAYISTPFHGSTLSAFETPNEFASQFSLKSVAESVTEAAYCCLCCKKKKKKSEKESSKFFDLSLLTDPAYLVVLLSSGAVAISVTIFIINLPSHAENLGFSKARGAYLLSTVSTLDLVGRIGGSTLCDLNLIPKWCYFVGGLIFAGISLIILPFMVSYTWISVVCAFYGLASGVNNSVTTLVLAEILGVERLMSSFGISLFVNGILQLIGPPLGGLWFERDQNYFHMFIALGFIFLFGASLWLFMPFINRHKNKKKLMNDKLTTGKLEPFAEEEAEEVQDERNPKVVFTPEETPSQMNGSPPMFKNKSNSNFEDATSPSKDFARSASAIQVAQNNERFRKVSTPASSRHGLQNISSRSHLPSTPSLLGSVPEGKASRVNSQEAFGKKLAIPKTPKRSPSTSSFQYMSTPFHGSTLSAFEKPSEFASQFSLKSVTDSLAPISYCCGCKKKEDDPKKEPNRYFDVQLLKDPIYLVILISNCTCAISYTNFIILVPAYANECGFDKSLGAYLLSIISALDLVGRIGGSALSDVVSTPKRYFFIFGLLLSGVSLSLIPLVRSYSAISAFCSIFGIASGINVGVTALVMTEMLGTERLMSSYGISLFMNGILQLIGPPICGFWFEYAKSYKSLFLTFGLILIGGAALWGLVPIINRRRRLAEQNKQTVGKA
Summary
Uniprot
Pubmed
Proteomes
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
Ontologies
GO
Topology
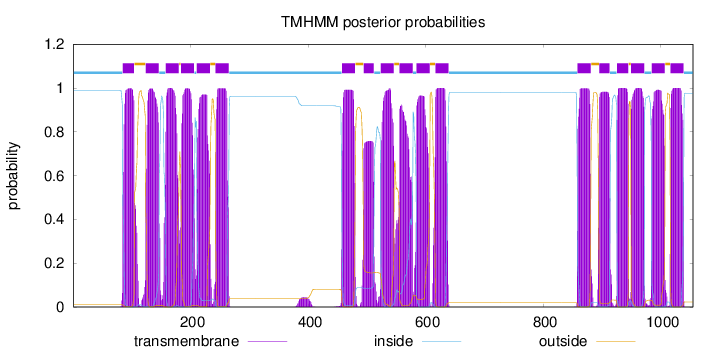
Length:
1055
Number of predicted TMHs:
18
Exp number of AAs in TMHs:
385.64937
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98913
inside
1 - 84
TMhelix
85 - 104
outside
105 - 123
TMhelix
124 - 146
inside
147 - 157
TMhelix
158 - 180
outside
181 - 183
TMhelix
184 - 206
inside
207 - 210
TMhelix
211 - 233
outside
234 - 242
TMhelix
243 - 265
inside
266 - 457
TMhelix
458 - 480
outside
481 - 494
TMhelix
495 - 512
inside
513 - 523
TMhelix
524 - 546
outside
547 - 555
TMhelix
556 - 578
inside
579 - 584
TMhelix
585 - 607
outside
608 - 616
TMhelix
617 - 639
inside
640 - 858
TMhelix
859 - 881
outside
882 - 895
TMhelix
896 - 913
inside
914 - 925
TMhelix
926 - 945
outside
946 - 949
TMhelix
950 - 972
inside
973 - 984
TMhelix
985 - 1007
outside
1008 - 1016
TMhelix
1017 - 1039
inside
1040 - 1055
Population Genetic Test Statistics
Pi
235.139404
Theta
181.07713
Tajima's D
0.97045
CLR
0.460079
CSRT
0.646417679116044
Interpretation
Uncertain
